EF hand
| EF hand | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | efhand | ||||||||
| Pfam | PF00036 | ||||||||
| Pfam clan | CL0220 | ||||||||
| ECOD | 108.1.1 | ||||||||
| InterPro | IPR002048 | ||||||||
| PROSITE | PDOC00018 | ||||||||
| SCOP2 | 1osa / SCOPe / SUPFAM | ||||||||
| CDD | cd00051 | ||||||||
| |||||||||
The EF hand is a helix–loop–helix structural domain or motif found in a large family of calcium-binding proteins.
The EF-hand motif contains a helix–loop–helix topology, much like the spread thumb and forefinger of the human hand, in which the Ca2+ ions are coordinated by ligands within the loop. The motif takes its name from traditional nomenclature used in describing the protein parvalbumin, which contains three such motifs and is probably involved in muscle relaxation via its calcium-binding activity.
The EF-hand consists of two alpha helices linked by a short loop region (usually about 12 amino acids) that usually binds calcium ions. EF-hands also appear in each structural domain of the signaling protein calmodulin and in the muscle protein troponin-C.
Calcium ion binding site
[edit]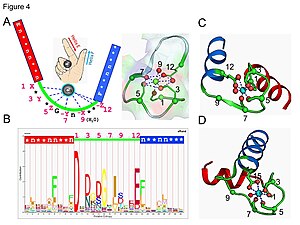
The calcium ion is coordinated in a pentagonal bipyramidal configuration. The six residues involved in the binding are in positions 1, 3, 5, 7, 9 and 12; these residues are denoted by X, Y, Z, -Y, -X and -Z. The invariant Glu or Asp at position 12 provides two oxygens for liganding calcium (bidentate ligand).
The calcium ion is bound by both protein backbone atoms and by amino acid side chains, specifically those of the anionic amino acid residues aspartate and glutamate. These residues are negatively charged and will make a charge-interaction with the positively charged calcium ion. The EF hand motif was among the first structural motifs whose sequence requirements were analyzed in detail. Five of the loop residues bind calcium and thus have a strong preference for oxygen-containing side chains, especially aspartate and glutamate. The sixth residue in the loop is necessarily glycine due to the conformational requirements of the backbone. The remaining residues are typically hydrophobic and form a hydrophobic core that binds and stabilizes the two helices.
Upon binding to Ca2+, this motif may undergo conformational changes that enable Ca2+-regulated functions as seen in Ca2+ effectors such as calmodulin (CaM) and troponin C (TnC) and Ca2+ buffers such as calreticulin and calbindin D9k. While the majority of the known EF-hand calcium-binding proteins (CaBPs) contain paired EF-hand motifs, CaBPs with single EF hands have also been discovered in both bacteria and eukaryotes. In addition, "EF-hand-like motifs" have been found in a number of bacteria. Although the coordination properties remain similar with the canonical 29-residue helix–loop–helix EF-hand motif, the EF-hand-like motifs differ from EF-hands in that they contain deviations in the secondary structure of the flanking sequences and/or variation in the length of the Ca2+-coordinating loop.
EF hands have very high selectivity for calcium. For example, the dissociation constant of alpha parvalbumin for Ca2+ is ~1000 times lower than that for the similar ion Mg2+.[2] This high selectivity is due to the relatively rigid coordination geometry, the presence of multiple charged amino acid side chains in the binding site, as well as the ion solvation properties.[3][4][5]
Prediction
[edit]
Pattern (motif signature) search is one of the most straightforward ways to predict continuous EF-hand Ca2+-binding sites in proteins. Based on the sequence alignment results of canonical EF-hand motifs, especially the conserved side chains directly involved in Ca2+ binding, a pattern PS50222 has been generated to predict canonical EF-hand sites. Prediction servers may be found in the external links section.
Classification
[edit]Since the delineation of the EF-hand motif in 1973, the family of EF-hand proteins has expanded to include at least 66 subfamilies thus far. EF-hand motifs are divided into two major structural groups:
- Canonical EF-hands as seen in calmodulin (CaM) and the prokaryotic CaM-like protein calerythrin. The 12-residue canonical EF-hand loop binds Ca2+ mainly via sidechain carboxylates or carbonyls (loop sequence positions 1, 3, 5, 12). The residue at the –X axis coordinates the Ca2+ ion through a bridged water molecule. The EF-hand loop has a bidentate ligand (Glu or Asp) at axis –Z.
- Pseudo EF-hands exclusively found in the N-termini of S100 and S100-like proteins. The 14-residue pseudo EF-hand loop chelates Ca2+ primarily via backbone carbonyls (positions 1, 4, 6, 9).
Additional points:
- EF-hand-like proteins with diversified flanking structural elements around the Ca2+-binding loop have been reported in bacteria and viruses. These prokaryotic EF-hand-like proteins are widely implicated in Ca2+ signaling and homeostasis in bacteria. They contain flexible lengths of Ca2+-binding loops that differ from the EF-hand motifs. However, their coordination properties resemble classical EF-hand motifs.
- For example, the semi-continuous Ca2+-binding site in D-galactose-binding protein (GBP) contains a nine-residue loop. The Ca2+ ion is coordinated by seven protein oxygen atoms, five of which are from the loop mimicking the canonical EF-loop whereas the other two are from the carboxylate group of a distant Glu.
- Another example is a novel domain named Excalibur (extracellular Ca2+-binding region) isolated from Bacillus subtilis. This domain has a conserved 10-residue Ca2+-binding loop strikingly similar to the canonical 12-residue EF-hand loop.
- The diversity of the structure of the flanking region is illustrated by the discovery of EF-hand-like domains in bacterial proteins. For example, a helix–loop–strand instead of the helix–loop–helix structure is in periplasmic galactose-binding protein (Salmonella typhimurium, PDB: 1gcg) or alginate-binding protein (Sphingomonas sp., 1kwh); the entering helix is missing in protective antigen (Bacillus anthracis, 1acc) or dockerin (Clostridium thermocellum, 1daq).
Among all the structures reported to date, the majority of EF-hand motifs are paired either between two canonical or one pseudo and one canonical motifs. For proteins with odd numbers of EF-hands, such as the penta-EF-hand calpain, EF-hand motifs were coupled through homo- or hetero-dimerization. The recently-identified EF-hand containing ER Ca2+ sensor protein, stromal interaction molecule 1 and 2 (STIM1, STIM2), has been shown to contain a Ca2+-binding canonical EF-hand motif that pairs with an immediate, downstream atypical "hidden" non-Ca2+-binding EF-hand. Single EF-hand motifs can serve as protein-docking modules: for example, the single EF hand in the NKD1 and NKD2 proteins binds the Dishevelled (DVL1, DVL2, DVL3) proteins.
Functionally, the EF-hands can be divided into two classes:
- signaling proteins
- buffering/transport proteins.
The first group is the largest and includes the most well-known members of the family such as calmodulin, troponin C and S100B. These proteins typically undergo a calcium-dependent conformational change which opens a target binding site. The latter group is represented by calbindin D9k and these proteins do not undergo calcium dependent conformational changes.
Subfamilies
[edit]Examples
[edit]Aequorin
[edit]Aequorin is a calcium binding protein (CaBP) isolated from the cnidarian Aequorea victoria. Aequorin belongs to the EF-hand family of CaBPs, with EF-hand loops that are closely related to CaBPs in mammals. In addition, aequorin has been used for years as an indicator of Ca2+ and has been shown to be safe and well tolerated by cells. Aequorin is made up of two components – the calcium binding component apoaequorin (AQ) and the chemiluminescent molecule coelenterazine. The AQ portion of this protein contains the EF-hand calcium binding domains.[6]
Human proteins
[edit]Humans proteins containing this domain include:
- ACTN1; ACTN2; ACTN3; ACTN4; APBA2BP; AYTL1; AYTL2
- C14orf143; CABP1; CABP2; CABP3; CABP4; CABP5; CABP7; CALB1; CALB2; CALM2; CALM3; CALML3; CALML4; CALML5; CALML6; CALN1; CALU; CAPN1; CAPN11; CAPN2; CAPN3; CAPN9; CAPNS1; CAPNS2; CAPS; CAPS2; CAPSL; CBARA1; CETN1; CETN2; CETN3; CHP; CHP2; CIB1; CIB2; CIB3; CIB4; CRNN
- DGKA; DGKB; DGKG; DST; DUOX1; DUOX2
- EFCAB1; EFCAB2; EFCAB4A; EFCAB4B; EFCAB6; EFCBP1; EFCBP2; EFHA1; EFHA2; EFHB; EFHC1; EFHD1; EFHD2; EPS15; EPS15L1
- FKBP10; FKBP14; FKBP7; FKBP9; FKBP9L; FREQ; FSTL1; FSTL5
- GCA; GPD2; GUCA1A; GUCA1B; GUCA1C
- hippocalcin; HPCAL1; HPCAL4; HZGJ
- IFPS; ITSN1; ITSN2; KCNIP1; KCNIP2; KCNIP3; KCNIP4; KIAA1799
- LCP1
- MACF1; MRLC2; MRLC3; MST133; MYL1; MYL2; MYL5; MYL6B; MYL7; MYL9; MYLC2PL; MYLPF
- NCALD; NIN; NKD1; NKD2; NLP; NOX5; NUCB1; NUCB2
- OCM
- PDCD6; PEF1; PKD2; PLCD1; PLCD4; PLCH1; PLCH2; PLS1; PLS3; PP1187; PPEF1; PPEF2; PPP3R1; PPP3R2; PRKCSH; PVALB
- RAB11FIP3; RASEF; RASGRP; RASGRP1; RASGRP2; RASGRP3; RCN1; RCN2; RCN3; RCV1; RCVRN; REPS1; RHBDL3; RHOT1; RHOT2; RPTN; RYR2; RYR3
- S100A1; S100A11; S100A12; S100A6; S100A8; S100A9; S100B; S100G; S100Z; SCAMC-2; SCGN; SCN5A; SDF4; SLC25A12; SLC25A13; SLC25A23; SLC25A24; SLC25A25; SPATA21; SPTA1; SPTAN1; SRI
- TBC1D9; TBC1D9B; TCHH; TESC; TNNC1; TNNC2
- USP32
- VSNL1
- ZZEF1
See also
[edit]- Another distinct calcium-binding motif composed of alpha helices is the dockerin domain.
References
[edit]- ^ Ban C, Ramakrishnan B, Ling KY, Kung C, Sundaralingam M (January 1994). "Structure of the recombinant Paramecium tetraurelia calmodulin at 1.68 A resolution". Acta Crystallogr. D. 50 (Pt 1): 50–63. doi:10.1107/S0907444993007991. PMID 15299476.
- ^ Schwaller, B. (13 October 2010). "Cytosolic Ca2+ Buffers". Cold Spring Harbor Perspectives in Biology. 2 (11): a004051. doi:10.1101/cshperspect.a004051. PMC 2964180. PMID 20943758.
- ^ Gifford, Jessica L.; Walsh, Michael P.; Vogel, Hans J. (15 July 2007). "Structures and metal-ion-binding properties of the Ca -binding helix–loop–helix EF-hand motifs". Biochemical Journal. 405 (2): 199–221. doi:10.1042/BJ20070255. PMID 17590154.
- ^ Dudev, Todor; Lim, Carmay (16 September 2013). "Competition among Metal Ions for Protein Binding Sites: Determinants of Metal Ion Selectivity in Proteins". Chemical Reviews. 114 (1): 538–556. doi:10.1021/cr4004665. PMID 24040963.
- ^ Jing, Zhifeng; Liu, Chengwen; Qi, Rui; Ren, Pengyu (23 July 2018). "Many-body effect determines the selectivity for Ca and Mg in proteins". Proceedings of the National Academy of Sciences. 115 (32): E7495–E7501. doi:10.1073/pnas.1805049115. PMC 6094099. PMID 30038003.
- ^ Detert JA, Adams EL, Lescher JD, Lyons JA, Moyer JR (2013). "Pretreatment with Apoaequorin Protects Hippocampal CA1 Neurons from Oxygen-Glucose Deprivation". PLOS ONE. 8 (11): e79002. doi:10.1371/journal.pone.0079002. PMC 3823939. PMID 24244400.
Further reading
[edit]- Branden C, Tooze J (1999). "Chapter 2: Motifs of protein structure". Introduction to Protein Structure. New York: Garland Pub. pp. 24–25. ISBN 0-8153-2305-0.
- Nakayama S, Kretsinger RH (1994). "Evolution of the EF-hand family of proteins". Annu Rev Biophys Biomol Struct. 23: 473–507. doi:10.1146/annurev.bb.23.060194.002353. PMID 7919790.
- Zhou Y, Yang W, Kirberger M, Lee HW, Ayalasomayajula G, Yang JJ (November 2006). "Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins". Proteins. 65 (3): 643–55. doi:10.1002/prot.21139. PMID 16981205. S2CID 8904181.
- Zhou Y, Frey TK, Yang JJ (July 2009). "Viral calciomics: interplays between Ca2+ and virus". Cell Calcium. 46 (1): 1–17. doi:10.1016/j.ceca.2009.05.005. PMC 3449087. PMID 19535138.
- Nakayama S, Moncrief ND, Kretsinger RH (May 1992). "Evolution of EF-hand calcium-modulated proteins. II. Domains of several subfamilies have diverse evolutionary histories". J. Mol. Evol. 34 (5): 416–48. doi:10.1007/BF00162998. PMID 1602495. S2CID 34614223.
- Hogue CW, MacManus JP, Banville D, Szabo AG (July 1992). "Comparison of terbium (III) luminescence enhancement in mutants of EF hand calcium binding proteins". J. Biol. Chem. 267 (19): 13340–7. doi:10.1016/S0021-9258(18)42216-8. PMID 1618836.
- Bairoch A, Cox JA (September 1990). "EF-hand motifs in inositol phospholipid-specific phospholipase C". FEBS Lett. 269 (2): 454–6. doi:10.1016/0014-5793(90)81214-9. PMID 2401372.
- Finn BE, Forsén S (January 1995). "The evolving model of calmodulin structure, function and activation". Structure. 3 (1): 7–11. doi:10.1016/S0969-2126(01)00130-7. PMID 7743133.
- Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M (October 2008). "Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry". Cell. 135 (1): 110–22. doi:10.1016/j.cell.2008.08.006. PMID 18854159.
- Nelson MR, Thulin E, Fagan PA, Forsén S, Chazin WJ (February 2002). "The EF-hand domain: a globally cooperative structural unit". Protein Sci. 11 (2): 198–205. doi:10.1110/ps.33302. PMC 2373453. PMID 11790829.
External links
[edit]- Eukaryotic Linear Motif resource motif class LIG_EH_1
- Eukaryotic Linear Motif resource motif class LIG_IQ
- Eukaryotic Linear Motif resource motif class DOC_PP2B_LxvP_1
- Eukaryotic Linear Motif resource motif class LIG_IQ
- Nelson M, Chazin W. "EF-Hand Calcium-Binding Proteins Data Library". Vanderbilt University. Retrieved 2009-08-29.
- Haiech J. "EF-hand protein database (EF-handome)". European Calcium Society and the Université Libre de Bruxelles. Retrieved 2009-08-29.
upon request to haiech@pharma.u-strasbg.fr
- Yang J. "Calciomics". Georgia State University. Archived from the original on 2009-10-12. Retrieved 2009-08-29.
prediction server for EF-hand calcium binding proteins
