Velvet complex
Velvet complex is a group of proteins found in fungi and especially molds that are important in reproduction and production of secondary metabolites including penicillin. The core members of the complex include VeA, LaeA (loss of aflR-expression A), and VelB.[1][2][3] Other proteins including VelC and VosA sometimes function in the complex.[1] The proteins were first characterized in Aspergillus nidulans.[2]
Some proteins in the complex are light-sensitive, including the founding member, VeA (Velvet A), which was first described in 1965.[2]
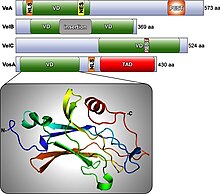
Four of these proteins, VeA, VelB, VelC, and VosA, have an approximately 200 amino acid domain called the velvet domain.[2]
Some fungal infections that are present in humans and sometimes plants have been traced down to certain velvet complex elements.
The Velvet complex seems to affect a number of functions that are of pathogenic nature. This process is facilitated, managed and administered by the proteins of velvet complex.[4]
Velvet complex proteins have also been encountered in the genome sequence of fungal organisms in the form of transcription factors.[5]
References
[edit]- ^ a b Martín, JF (May 2017). "Key role of LaeA and velvet complex proteins on expression of β-lactam and PR-toxin genes in Penicillium chrysogenum: cross-talk regulation of secondary metabolite pathways". Journal of Industrial Microbiology & Biotechnology. 44 (4–5): 525–535. doi:10.1007/s10295-016-1830-y. PMID 27565675. S2CID 20855430.
- ^ a b c d e Gerke, J; Braus, GH (October 2014). "Manipulation of fungal development as source of novel secondary metabolites for biotechnology". Applied Microbiology and Biotechnology. 98 (20): 8443–55. doi:10.1007/s00253-014-5997-8. PMC 4192562. PMID 25142695.
- ^ Amare, MG; Keller, NP (May 2014). "Molecular mechanisms of Aspergillus flavus secondary metabolism and development". Fungal Genetics and Biology. 66: 11–8. doi:10.1016/j.fgb.2014.02.008. PMID 24613992.
- ^ López-Berges, Manuel S.; Hera, Concepción; Sulyok, Michael; Schäfer, Katja; Capilla, Javier; Guarro, Josep; Di Pietro, Antonio (2012-11-19). "The velvet complex governs mycotoxin production and virulence ofFusarium oxysporumon plant and mammalian hosts". Molecular Microbiology. 87 (1): 49–65. doi:10.1111/mmi.12082. ISSN 0950-382X. PMID 23106229. S2CID 6237805.
- ^ Wang, Rui; Leng, Yueqiang; Shrestha, Subidhya; Zhong, Shaobin (August 2016). "Coordinated and independent functions of velvet-complex genes in fungal development and virulence of the fungal cereal pathogen Cochliobolus sativus". Fungal Biology. 120 (8): 948–960. Bibcode:2016FunB..120..948W. doi:10.1016/j.funbio.2016.05.004. ISSN 1878-6146. PMID 27521627.
