Talk:Genetic history of the Indigenous peoples of the Americas
| This is the talk page for discussing improvements to the Genetic history of the Indigenous peoples of the Americas article. This is not a forum for general discussion of the article's subject. |
Article policies
|
| Find sources: Google (books · news · scholar · free images · WP refs) · FENS · JSTOR · TWL |
| This article is rated B-class on Wikipedia's content assessment scale. It is of interest to the following WikiProjects: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toolbox |
|---|
Nomenclature used in this article
[edit]According to the YCC guidelines proposed in 2002, and used in most journals now, there are two options - either Q1a3a (phylogenetic name) or else Q-M3 (mutational name). So Q1a3a-M3 or Q1a3aM3 do not exist. They both look like they would possibly mean something else, like for example the name of an STR derived cluster within the clade. Wikipedia should also eventually standardize.--Andrew Lancaster (talk) 16:04, 1 December 2009 (UTC)
 Done ok edited... i think pls double check ..I have added your definitions to the article ..i also added (may be too technical for most readers to understand.) template ....Buzzzsherman (talk) 22:48, 9 December 2009 (UTC)
Done ok edited... i think pls double check ..I have added your definitions to the article ..i also added (may be too technical for most readers to understand.) template ....Buzzzsherman (talk) 22:48, 9 December 2009 (UTC)
- Removed {{technical}}..I put it there ..but i think all can understated now ..many copy edits later :) ..!!!.....Buzzzsherman (talk) 06:00, 27 January 2010 (UTC)
Readability
[edit]I'd say the first paragraph of the introduction could be reworded to be more...introductory. Also, with an image that big right at the start, the text gets pushed out of the way (down to literally one word per line on my screen). The image wouldn't be effective at a small enough size...maybe put it later in the article? The "Y-DNA Q haplogroup tree" could definitely be organized more clearly, but I'm not sure how you could go about that. That section also has some grammatical errors that make understanding more difficult. As written, the article will definitely present a challenge to the average reader. Cheers, Nikkimaria (talk) 13:55, 30 January 2010 (UTC)
- I love you TKS!!!... Think i will crop the first image!! ...ok i will work on every point...The into I agree not my strong suit...grammatical errors dame i always have this problem. Buzzzsherman (talk) 18:35, 30 January 2010 (UTC)
- I have reduced the image size and moved it.. cropping is not working for me :(....re did some of that tree... like you not sure who to show it better...AS for intro i think i will ask one of the WikiProject Human Genetic History guys to have a look! My problem is the title of the article and trying to incorporate that into the first sentences..I have already renamed it 2 times :) .....Buzzzsherman (talk) 18:55, 30 January 2010 (UTC)
- Update i redid the intro a bit... since we talk about Autosomal DNA now..so i hope more legible...Buzzzsherman (talk) 00:40, 3 February 2010 (UTC)
Autosomal DNA
[edit]The article currently only includes Y-DNA and mtDNA information. But these are just two markers out of many. I would suggest include some information from autosomal DNA markers. Cavalli-Sforza's History and Geography of Human Genes may be a good place to start. Wapondaponda (talk) 08:22, 31 January 2010 (UTC)
- I was going to mention it ..but since its not really used whaen talking about Paleo-Indians, Paleo-Eskimos etc..More so in direct family genetics and also, autosomal markers contain all your health issues. ..I dont want to confuse anyone..But in time i think it will be added as methods get better!!
- This bellow is the typical stuff i find on the subject.. However, pls if anyone can help add this subject to the article DO SO!!!!!
| “ | The ONLY testing today that can positively identify Native American or African ancestry is the Y-chromosome and mitochondrial DNA testing. This is due to the stability of the markers used for these tests to determine the haplogroup (your twig on the World Family Tree) for the basic ethnic groups. Testing particular parts of the Y-chromosome and the mitochondrial markers are reliable as they are consistent over time. That is to say, they rarely mutate, and those minor mutations help geneticists and genetic genealogists group testers into families. Geneticists have determined which twigs on the World Family Tree (Phylogenetic Tree) do tend to have Western European, Asian, Native American and African DNA; therefore, by testing either the all male line (Ydna test) or the all female line (mtDNA test) an accurate determination of these cultural groups can be made.
Some testing companies give a percentage for various ethnic groups and claim to assign a tribe to African or Native American Heritage with atDNA. The test result from these companies is displayed as percentages for various ethnic groups. Not every ethnic group can be determined. Basically, a tester would get a percentage for Western European, Native American, African, and Asian. For example, a person’s test could result in 36% Western European, 24% African, etc. A genealogist would have to ask how helpful a percentage of a cultural group is to the search for ancestors. "Autosomal markers are best used to help determine paternity and for use in forensics". Emily Aulicino a Genetic Genealogist for Genealogical Forum of Oregon, Inc. |
” |
- -
Buzzzsherman (talk) 05:46, 2 February 2010 (UTC)- The strength of mtDNA and y-chromosome markers is that they can be used to establish phylogenetic/genealogical relationships between individuals. However, their weakness is that they are not as useful as autosomal markers for studying phenotypes since only a few mtDNA and y-chromosome markers are used, and most markers are selectively neutral("junk DNA"). Since the article only includes mtDNA and y-chromosome information, then a more appropriate name would be "Indigenous American haplogroups", or "Indigenous American genetic genealogy". Genetics is a much broader field that relates to the genome as a whole, including mtDNA, y-chromosome and the autosomes. From a biomedical perspective, autosomal markers are more useful, as they can be used to diagnose, treat or prevent disease. But I do agree that autosomal markers overlap significantly, so much so that few, if any, are unique to any given population. Nonetheless populations do differ in frequencies of DNA variants and this information is useful in biomedicine. For example, Indigenous Americans almost exclusively belong to blood group O.
- Another feature of Native American genetics, which applies to y-DNA, mtDNA and the autosomes, is a lower level of genetic diversity relative to other continental populations, supporting the theory that the the Americas were the last continents to be settled. Furthermore, genetic diversity decreases in a southward direction supporting the hypothesis that the Americas were settled from the north. Finally Native Americans genetics are a subset of siberian genetics consistent with a Siberian origin of Native Americans. As this information applies to all types of markers I believe it should be included in any general article about native American genetics. A useful article is,
- Genetic Variation and Population Structure in Native Americans
Wapondaponda (talk) 18:06, 2 February 2010 (UTC)
- Ok i like your argument ...
but i dont feel i am knowledgeable enough to add atDNA info...So lets move the page to Indigenous Amerindian haplogroups..We(Wikipedia:WikiProject Indigenous peoples of North America have chosen not to use the word America or Native long ago.. as it may be misleading as most might think it is only for the USA.. PS i see your from the Genetics project...thank you very very much in helping ...Not to many have the knowledge to help here TKS MAN!!!
- Ok i like your argument ...
...damit the more i read your link the more i see that it should be added!!..ok give me some time will read a book or 2 then add some info..pls fell free anyone to jumpin and help!!! ...Buzzzsherman (talk) 20:05, 2 February 2010 (UTC)
- UPDATE "Why Strike above" ok i have added a section..basically a copy edit of this (All site content, except where otherwise noted, is licensed under a Creative Commons Attribution License.) ..I will add more as i read more ..Buzzzsherman (talk) 22:12, 2 February 2010 (UTC)
- Here are some observations concerning the article,
- The lead states,
- "Indigenous Amerindian genetics primarily focus on Human Y-chromosome DNA haplogroups and Human mitochondrial DNA haplogroups."
- Genealogy is one of the most popular hobbies in the western world. MtDNA and Y-DNA are powerful tools for determining ancient genealogical relationships. It is therefore no surprise that public interest in y-DNA and mtDNA has exploded since the discovery of a mitochondrial Eve in 1987. It is good for science that regular people have taken a keen interest in mtDNA and Y-DNA studies. However such interest is not without its problems and controversies. There is the potential for misunderstanding what they mean, and in some cases there has been politicization of DNA information [1], [2]. Y-DNA and mtDNA tell stories that no other DNA marker can, but y-DNA and mtDNA markers only represent a tiny fraction of human genetic variability. Consequently I think it is somewhat inaccurate for Indigenous Amerindian genetics to be primarily about Y-DNA and mtDNA, though these markers are very popular in the public's eye.
- UPDATE "Why Strike above" ok i have added a section..basically a copy edit of this (All site content, except where otherwise noted, is licensed under a Creative Commons Attribution License.) ..I will add more as i read more ..Buzzzsherman (talk) 22:12, 2 February 2010 (UTC)
- What is interesting about Amerindian genetics is that autosomal, y-DNA and mtDNA all display a similar pattern. All three types all share common ancestry with Siberian/East Asian populations. All three types display a loss of diversity as one moves south. IOW y-DNA, mtDNA and autosomal haplotypes in South America are subclades of North American haplotypes. As this pattern is applicable to the genome as a whole, I would suggest having a summary of this similarity in the lead.
- Many wikipedia articles under WP:HGH use the word "genetics" in their title, but what they are actually referring to is population genetics and not the genetics of individuals. This distinction is important, because save for twins/clones, all individuals are genetically unique. This article is actually referring to "Indigenous American population genetics". :::Wapondaponda (talk) 17:33, 3 February 2010 (UTC)
- Ok what are you saying ?...what should we change in the lead?...and should we move the article to Indigenous Amerindian population genetics???......As for what the public uses i cant help that... only state that they do use Y dna more!!..Buzzzsherman (talk) 19:12, 3 February 2010 (UTC)
- There are many ways to approach the subject matter. The article Genetic history of Europe has focused on what genes reveal about the history and pre-history of Europe. Consequently Y-DNA and mtDNA can play a prominent role as the strength of these markers is establishing patterns of prehistoric migrations. The article, Genetics and archeogenetics of South Asia has taken another approach in its title, though the science used is similar to that found in genetic history of Europe.
- I think it is important to establish the scope of the article, and the scope of the article should be consistent with the title of the article. Based on the title "Indigenous Amerindian genetics", my initial impression was that the scope of the article was indeed "Indigenous American population genetics". If this is the case, atDNA should be covered, in addition to Y-DNA and mtDNA, but not just as a step-child. While Y-DNA and mtDNA have been studied in detail recently, much of the initial work in population genetics was based on polymorphisms found in blood proteins, which are coded by autosomal DNA. So there is a substantial body of work on atDNA markers as well.
- Some general DNA studies include
- Cavalli-Sforza (1994). "America". History and Geography of Human Genes.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - Mulligan; et al. (2004). "Population genetics, history, and health patterns in native americans".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - Bourgeois; et al. (2009). "X-chromosome lineages and the settlement of the Americas".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - Mesa; et al. (2000). "Autosomal, mtDNA, and Y-Chromosome Diversity in Amerinds: Pre- and Post-Columbian Patterns of Gene Flow in South America".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - Estrada-Mena; et al. (2009). "Blood group O alleles in Native Americans: Implications in the peopling of the Americas".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help)
- Cavalli-Sforza (1994). "America". History and Geography of Human Genes.
- Wapondaponda (talk) 10:04, 4 February 2010 (UTC)
- Ok i will look this over...BUT PLS ADD WHAT YOU THINK IS NEEDED if you like ...I am the creator of the article ....but hope that others will help !!...Buzzzsherman (talk) 16:51, 4 February 2010 (UTC)
- Ok what are you saying ?...what should we change in the lead?...and should we move the article to Indigenous Amerindian population genetics???......As for what the public uses i cant help that... only state that they do use Y dna more!!..Buzzzsherman (talk) 19:12, 3 February 2010 (UTC)
Incorrect terminology. I enjoyed this article, but I believe the term 'cellular differentiation' as written in the following sentence of the 'atDNA' section, "The Amerindian populations show a lower genetic diversity and cellular differentiation than populations from other continental regions" is used incorrectly. This term is very specific to developmental biology and has nothing to do with the topic at hand (ie. genetic diversity of Amerindians.) The article for ref. 31 states, "...lower genetic diversity and greater differentiation than populations from other continental regions." Thus, I have edited the article. FWIW, I wonder if these two concepts (1. genetic diversity - the variation within a gene pool and 2. differentiation - degree of difference between gene pools) are too difficult for lay people to grasp. Also, ref. 81 is unrelated to the topic and perhaps ought to be removed. Das312 (talk) 13:47, 15 May 2010 (UTC)
- Hello there, i am glad you enjoyed reading the article...Yes if you think something is odd/or misleading pls change it....as for ref #81 looks like it got move in a copy edit ..i will look it over soon see were it goes....Moxy (talk) 18:13, 15 May 2010 (UTC)
10
[edit]A factor of 10 is mentioned in the article. There is no time scale mentioned. —Preceding unsigned comment added by 81.148.18.65 (talk) 12:53, 24 May 2010 (UTC)
 Fixed and then they grew by a factor of 10 over 800 – 1000 yearsWells, Spencer; Read, Mark (2002) "The Journey of Man" - A Genetic Odyssey. Random House. pp. 138–140.. Moxy (talk) 20:36, 20 December 2010 (UTC)
Fixed and then they grew by a factor of 10 over 800 – 1000 yearsWells, Spencer; Read, Mark (2002) "The Journey of Man" - A Genetic Odyssey. Random House. pp. 138–140.. Moxy (talk) 20:36, 20 December 2010 (UTC)
Specific european y-chromosome haplogroups
[edit]Does anyone know the specific european y-chromosome haplogroups that native american males have? —Preceding unsigned comment added by 68.238.6.118 (talk) 23:29, 24 February 2011 (UTC)
- Native northamerican males have mainly R1b and R1a, but the available information is very poor. --Mauricio (talk) 01:16, 3 April 2011 (UTC)
MtDNA section
[edit]This section has information on haplogroup X that seems contradictory and may need a little clarification. I've read it several times and can't get my head around it.
Haplogroup X (mtDNA) is one of the haplogroups associated with the single founding east Asian population that initially populated the Americas; how can it also be not at all strongly associated with east Asia? I come up with two possibilites, which is why I thought it may need clarification. Did Haplogroup X enter the east Asian population 20-30,000 years ago, or did a group from the Altai region, somehow travel to and get stuck on Beringia along with the other east Asian populations?
I know, fussy work. Great article though. Nihola (talk) 16:30, 29 March 2011 (UTC)
- The single founding that initially populated the Americas is Not associated with the East Asian population, but Siberian population. For its part, in its time, Siberia was initially populated by east asian haplogroups A, B, C and D, and the west (or central) asian X.--Mauricio (talk) 19:01, 10 April 2011 (UTC)
Mitochondrial haplogroup M discovered in prehistoric North Americans
[edit]Mathilda’s Anthropology Blog. Just another WordPress.com weblog Skip to content HOME ABOUT ME AND MY BLOG
← European mt DNA U5 in a Chinese tomb.Mitochondrial DNA haplogroups and age at onset of schizophrenia → Mitochondrial haplogroup M discovered in prehistoric North Americans Posted on November 3, 2008 by mathilda37| Leave a comment
Mitochondrial haplogroup M discovered in prehistoric North Americans
Received 21 April 2006; received in revised form 8 July 2006; accepted 11 July 2006
We analyzed two mid-Holocene (w5000 years before present) individuals from North America who belong to mitochondrial DNA (mtDNA) haplogroup M, a common type found in East Asia, but one that has never before been reported in ancient or living indigenous populations in the Americas. This study provides evidence that the founding migrants of the Americas exhibited greater genetic diversity than previously recognized, prompting us to reconsider the widely accepted five-founder model that posits that the Americas were colonized by only five founding mtDNA lineages. Additional genetic studies of prehistoric remains in the Americas are likely to reveal important insights into the early population history of Native Americans. However, the usefulness of this information will be tempered by the ability of researchers to distinguish novel founding lineages from contamination and, as such, we recommend strategies to successfully accomplish this goal.
I’m surprised I’ve not heard more noise around this. Unfortunately I couldn’t spot any information in the paper on what known population this M clade this might be closest too.
In this study we demonstrate the existence of this undocumented genetic structure with the discovery of two individuals from China Lake, British Columbia (Fig. 1) that exhibit a mitochondrial DNA (mtDNA) haplogroup never before reported in any prehistoric or living indigenous population in the Americas. Both were found in the same burial, dated to 4950 +/- 170 14C years before present (ybp), and were believed to be related due to similar morphological characteristics (Cybulski personal communication). Both individuals belong to haplogroup M, which is widely distributed throughout Asia (Kivisild et al., 2002). These individuals account for two of three samples dating to approximately 5000 ybp studied from the Northern Plateau region in Northwestern North America that were compared to 3658 sequences from Native Americans widely distributed throughout the Americas. The third sample dating to 4975 � 40 14C ybp from Big Bar Lake, British Columbia (Fig. 1), exhibits a haplogroup A haplotype that is shared with contemporary indigenous individuals. The discovery of a new mitochondrial haplogroup in the Americas conflicts with the presumed five-founder model, which implies that all Native American mtDNA derives from only five lineages, the founding haplotypes of haplogroups A, B, C, D, and X (Eshleman et al., 2003). Our discovery demonstrates that a more genetically diverse group of migrants colonized the Americas than previously thought and supports the hypothesis that significant undocumented genetic diversity likely still remains in the Americas.
This now brings the founding American mt lineages to six; A, B, C, D, X and M.
Old world genetic admixture
[edit]In the "triangle plot" image, made from the cited study, only one group was identified at its point of origin - Hispanics, from a sample in southern Colorado. It seems that other groups should also be identified (i.e. African Americans from Washington, DC), or perhaps none at all?Parkwells (talk) 23:47, 21 September 2011 (UTC)
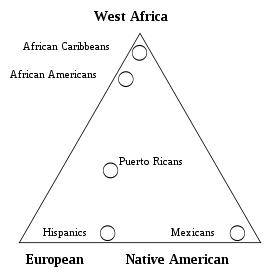
- On one hand, this is a great diagram that it should be somewhere. I don't agree after discussing scientific genetics that "self-identification" should be here. Seems like mixing myth with science. I suggest it be moved "elsewhere," but not sure where "elsewhere" should be. Somewhere where "self-identification" wouldn't seem unscientific! Student7 (talk) 18:18, 25 September 2011 (UTC)
- Student7, what world are you living in where Puerto Rican, Mexican, etc are only "self identifications"? --ಠ_ಠ node.ue ಠ_ಠ (talk) 23:33, 28 September 2011 (UTC)
- Please avoid WP:PERSONAL attacks. Thanks.
- It appears to me that "Mexicans" are the only group that might be germane to this particular topic. The other groups seem non-WP:TOPIC.Student7 (talk) 21:04, 29 September 2011 (UTC)
- Student7, what world are you living in where Puerto Rican, Mexican, etc are only "self identifications"? --ಠ_ಠ node.ue ಠ_ಠ (talk) 23:33, 28 September 2011 (UTC)
- I agree that the diagram does not relate to this article, which is about the origins of the indigenous peoples and their genetic history.Parkwells (talk) 22:28, 29 September 2011 (UTC)
Why is the term "race" being used in reference to genetics. Race is an imprecise social/cultural concept not a scientific term. — Preceding unsigned comment added by 76.64.40.22 (talk) 01:09, 22 December 2014 (UTC)
Genetic chart at the top
[edit]There is a great chart at the top. It only has two tribes on it from Canada and the US: Objibwa and Cree. Kind of invites the question: what happened to the those other tribes of NA? Student7 (talk) 21:04, 29 September 2011 (UTC)
- It relates to the languages as well - Na-Dene is the language group for Navajo and Apachean languages, among others; but the Navajo tribe is not listed on the chart. It's just a starting point. Parkwells (talk) 23:47, 29 September 2011 (UTC)
Error in the R1 section
[edit]The penultimate sentence in the R1 section beginning with "Principal component analysis..." is mistaken unless I'm seriously misunderstanding something. The Amerind groups with the highest proportion of type R1 (Chippewyan, etc.) listed in the R1 section do NOT "most closely resemble" the Siberians with the highest proportion of type Q (Ket, etc.) listed in the Q section. Of course, if I'm wrong, please cite the source of the principal component analysis that found that the American tribes with the highest proportion of R1 resemble the Siberians with the highest proportion of type Q more closely than they resemble any fellow R1s.
If I'm right, then I suspect that I'm not the only one who would love to know the real answer to the questions, Which non-American R1s do the Chippewyans and other majority R1 Eastern tribes most closely resemble, how closely, and why?
Tuanglen (talk) 21:39, 7 March 2012 (UTC)
- Its not non American - its relation is with mtDNA haplogroup X not Q YDNA. I hope this helpsMoxy (talk) 00:43, 8 March 2012 (UTC)
I apologize if there is some misunderstanding on my part. I notice that the sentence has been changed, but I still believe there is a problem. It now says,
"The principal-component analysis suggests a close genetic relatedness between some North American Amerindians (the Chipewyan and the Cheyenne) and certain populations of central/southern Siberia, at the resolution of major Y-chromosome haplogroups. This pattern agrees with the distribution of mtDNA haplogroup X...."
Which PCA suggests close relatedness between Chipewyan/Cheyenne and which populations of central/southern Siberia? Specifically. If an actual study were cited, I could read it myself, but none is. This is a section on Amerindian Y-DNA, so any mention of "the PCA suggests" must be assumed to be referring to a study of their Y-DNA. The text even says, "at the resolution of major Y-chromosome haplogroups." If the PCA analysis referred to in the section on the R1 Y-DNA of some Amerindians is not a PCA analysis of the R1 Y-DNA of those Amerindians, it ought to say what it actually is a study of, and cite the study.
Or else the claims about what this mysterious principal component analysis allegedly suggests or agrees with should be removed from the article. — Preceding unsigned comment added by Tuanglen (talk • contribs) 11:38, 8 March 2012 (UTC)
Tuanglen writes: "Which non-American R1s do the Chippewyans and other majority R1 Eastern tribes most closely resemble, how closely, and why?"
Closest STR matches are to French, English, and Scots. There is no evidence that the R1b in Native Americans originated from anything other than colonialism. — Preceding unsigned comment added by 24.77.173.247 (talk) 15:11, 20 June 2014 (UTC)
mtDNA haplogroups
[edit]You can only read the abstract on the new reference about C4c. But on the evidence available so far, the Amerindian haplogroups C4c and X2a have no demonstrated relationship to each other. While C4c has a clear Siberian origin, the X2a origin remains a mystery. It is not wise to take as valid a claim if it has not really been demonstrated.--Maulucioni (talk) 05:03, 30 March 2012 (UTC)
Book that makes suspicious claims of origins of indigenous Americans
[edit]I usually do not go to articles focused on genetics, but there was a small section in the book that didn't match up with what I was reading of the ancestry of the Amerindians. In the book: "La Florida: Spanish Exploration & Settlement of North America, 1500 to 1600", stating that Europeans might have sailed over from the Americas based on certain DNA markers found in certain Indian groups that are no more than 15,000 years ago, the Cactus Hill settlement, and that they couldn't have came from overland Asia b/c of a mile high glacier blocking the path. [3](here's the link). I just realized that it was the Solutrean hypothesis, and it is controversial b/c lack of peer reviews and what not. It doesn't match up with what I've read about the settlement of the Americas during the later Ice Age, and wondered what validity it has on being mentioned on this page. Please contact me as you can, and thanks for reading. LeftAire (talk) 15:17, 5 July 2012 (UTC)
- This is the author's bio[4]. Solitude Press seems to have published 16 books by him and only 2 or 3 by anyone else, so I suspect self-published. He's amateur and his books need to be read with care. Dougweller (talk) 16:34, 5 July 2012 (UTC)
I keep reading very tangled explanations of why Native Americans have such a high proportion of European genetic material, such as is found in this article. Bryan Sykes in DNA USA offers his estimate that (if memory serves) 20% to 25% of indigenous American genes are European in origin. Rather than just saying "dang, there sure was a lot of interbreeding among European settlers and native Americans," shouldn't the paragraph that addresses this issue at least mention the Solutrean hypothesis? I am just an amateur guesser, but I'm guessing that at some point during my lifetime ancient European settlement of the Americas will become the prevailing paradigm among scholars. — Preceding unsigned comment added by 2602:306:8B80:52E0:B5FB:5B9E:ABDD:5074 (talk) 01:50, 14 August 2015 (UTC)
- Native Americans and Europeans (and all kinds of other Old World immigrants) *did* intermarry a lot - this is amply documented. There is no mystery here. Of course, there could *also* be Solutrean ancestry, but the *proportion* of European ancestry is no evidence for it.
- Any admixture that *did* arrive through Solutreans will be easily distinguishable from modern European admixture when examined carefully, because they will have been separated for ~20 000 years. Mitochondrial haplogroup X2a is a popular candidate for Solutrean ancestry for just this reason - Europeans have a fair bit of X2, but it turns out to be an entirely different kind from Amerindian X2. (Not that there is any evidence that Solutreans had X2a: there is no known X in Europe before the Neolithic, the 2 known Solutreans had U and R0, but the sample size is too small to rule it out.) On the other hand R1b-M269, which is common in Amerindians and predominates in Western Europe, can't be Solutrean - it is much too young, and Amerindian R1b is just like European R1b when haplotypes are compared. I have yet to see any evidence of a Native American R1 clade, even though it wouldn't be surprising in principle - R1 is plenty old enough, very widespread, and related to Q.
- The pattern of admixture is also indicative of recent sources. For instance, if you look at the autosomal admixture graph from the recent study "Genomic evidence for Pleistocene and recent population history of Native Americans", you can see that within a given population, some individuals will have very high levels of European admixture, some will have low levels, and some will have none at all. This would not result from *ancient* admixture, because it would have all been homogenized to an even level long ago. What's more, there are actually two European components in this study, one which is most important in Europeans and minor in other people, and another which is actually highest in Middle Eastern populations: these two components appear in the same proportions in admixed Amerindians as they do in modern Europeans. Similarly, looking at the Y haplogroups in "Asymmetric male and female genetic histories among Native Americans from Eastern North America" and other studies, the proportion of R1 to other purportedly European haplogroups is the same in Native Americans (65%) as it is in European-Americans from Hammer et al 2005 (66%). The only real anomaly I am aware of in this respect is an often-studied sample of 48 Chipewyan men, which had 30 cases of R1 and no other European haplogroups.70.75.233.253 (talk) 18:46, 19 August 2015 (UTC)
Ancient Siberian genome reveals genetic origins of Native Americans
[edit]The study itself is at [5]. Another link is [6] and says "These findings may explain the presence of mitochondrial lineage X in Native Americans." and "the study concludes that two distinct Old World populations led to the formation of the First American gene pool: one related to modern-day East Asians, and the other a Siberian Upper Palaeolithic population related to modern-day western Eurasians." and "The presence of a population related to western Eurasians further into northeast Eurasia provides a more likely explanation for the presence of non-East Asian cranial characteristics in the First Americans, rather than the Solutrean hypothesis that proposes an Atlantic route from Iberia." Dougweller (talk) 19:10, 22 November 2013 (UTC)
- These full-genome studies are quite elusive. Non-recombining DNA studies are much better. YOMAL SIDOROFF-BIARMSKII (talk) 15:19, 9 March 2014 (UTC)
Hi, genetic history is not my forté. Would anyone familiar with the topic care to look at a recent addition Paiute people#Genetic history? I made it less sensationalist and linked back to this article but any further clean up would be greatly appreciated. -Uyvsdi (talk) 17:32, 4 February 2014 (UTC)Uyvsdi
- Just spotted "D-1 Haplogroup has also been found among people of Hokkaido, Japan, a northern region inhabited by ancient indigenous people known as the Ainu." The Ainu are a pretty modern ethnic group, first mentioned in the 13th century CE. The abstract (not a reliable source at times) itself says "However, probably due to the small sample size or close consanguinity among the members of the site, the frequencies of the haplogroups in Funadomari skeletons were quite different from any modern populations, including Hokkaido Ainu, who have been regarded as the direct descendant of the Hokkaido Jomon people." In any case, the second and third paragraph sources do not seem to mention the Paiute so should be removed. Dougweller (talk) 19:09, 4 February 2014 (UTC)
- A minor and somewhat tangential point, since it's not directly related to the Paiutes, but the first sentence did not originate from the abstract. The Ainu are regarded as Jōmon descendants and the Jōmon are indeed the most ancient identified cultural group in Japan. Strictly speaking, "Jōmon" would not be the correct term to use referring to a North American source population since it is a term referring to cultural markers that emerged after the migration was underway. "Jōmonoid" might be a better term to use for the ancestral population of the Jōmon culture and perhaps to the Ainu as well.75.111.54.141 (talk) 23:15, 26 April 2015 (UTC)
- Hi, have just started to work on this and was trying, first, to see what Kemp et al. did say. Sorry for the roughness by leaving material in that belongs elsewhere. Agree with you. Similarly, the criticism by Harry and her co-writer (last paragraph in that section) are about National Geographic's Genetic Project and has nothing to do directly with this material on the Paiute, so intend to delete that as well, and see if there is an article on National Geographic's project, where it belongs. Am reading the sources to see what they DO say. The link to the National Geographic Project is general, so may have difficulty tracking down what was stated.Parkwells (talk) 19:31, 4 February 2014 (UTC)
- There seems to be a new study making something like this connection: "During the deglaciation period, another group branched out from northern coastal China and traveled to Japan. "We were surprised to find that this ancestral source also contributed to the Japanese gene pool, especially the indigenous Ainus," says Li.
- This discovery helps to explain archeological similarities between the Paleolithic peoples of China, Japan, and the Americas"https://www.sciencedaily.com/releases/2023/05/230509122008.htm (edit to add: found a link to the actual article: https://www.cell.com/cell-reports/fulltext/S2211-1247(23)00424-2Johundhar (talk) 03:17, 10 May 2023 (UTC)
Solutrean hypothesis redux
[edit]I keep reading very tangled explanations of why Native Americans have such a high proportion of European genetic material, such as is found in this article. Bryan Sykes in DNA USA offers his estimate that (if memory serves) 20% to 25% of indigenous American genes are European in origin. Rather than just saying "dang, there sure was a lot of interbreeding among European settlers and native Americans," shouldn't the paragraph that addresses this issue at least mention the Solutrean hypothesis? I am just an amateur guesser, but I'm guessing that at some point during my lifetime ancient European settlement of the Americas will become the prevailing paradigm among scholars.104.184.5.46 (talk) 01:53, 14 August 2015 (UTC)
- The Solutrean Hypothesis cites Haplogroup X being found in high frequency among North Eastern Indigenous groups in Canada and the U.S., but what the hypothesis has failed to follow up on is that the X is actually X2a which has its origins in Central Asia, or Eurasia, going back 50,000 years ago. This X2a has been pinpointed as the progenitor to the subsequent X marker that is found in very low frequency in parts of Europe, but higher frequency in Asia Minor and the Levant, which are not considered European. Also, I have yet to see Dr Stanford or Bryan Sykes provide any genetic information on the actual Solutrean culture itself. As far as I know they still only have artefactual tools and artwork, so there is no common link to state that the Solutreans carried the X2a marker at all.--173.60.51.236 (talk) 18:44, 23 April 2016 (UTC)
- The "Solutrean hypothesis", while perfectly reasonable when first proposed (1980s?), has by now (after 2012 or so) been virtually ruled out. It seems now clear, as far as I have gathered, that there is certainly "European" genetic lineage found in Amerind lineages, but this isn't due to any early European trans-Atlantic contact, but it is perfectly explained by significant Ancient North Eurasian (ANE) expansion both into Beringia (and hence into America) and into Europe. So yes, the genetic relationship is there, but not because Amerinds are descended from Europeans, but because both Amerinds and Europeans are significantly descended from ANEs. --dab (𒁳) 10:44, 26 July 2018 (UTC)
External links modified
[edit]Hello fellow Wikipedians,
I have just modified 9 external links on Genetic history of indigenous peoples of the Americas. Please take a moment to review my edit. If you have any questions, or need the bot to ignore the links, or the page altogether, please visit this simple FaQ for additional information. I made the following changes:
- Added archive https://web.archive.org/web/20100622001311/http://www.genebase.com/tutorial/item.php?tuId=16 to http://www.genebase.com/tutorial/item.php?tuId=16
- Added archive https://web.archive.org/web/20121010092348/http://dsc.discovery.com/news/2008/02/13/beringia-native-american.html to http://dsc.discovery.com/news/2008/02/13/beringia-native-american.html
- Added archive https://web.archive.org/web/20120313061401/http://dsc.discovery.com/news/2008/02/13/beringia-native-american-02.html to http://dsc.discovery.com/news/2008/02/13/beringia-native-american-02.html
- Added archive https://web.archive.org/web/20100622001311/http://www.genebase.com/tutorial/item.php?tuId=16 to http://www.genebase.com/tutorial/item.php?tuId=16
- Added archive https://web.archive.org/web/20110511091253/https://www.cambridgedna.com/genealogy-dna-ancient-migrations-slideshow.php?view=step7 to https://www.cambridgedna.com/genealogy-dna-ancient-migrations-slideshow.php?view=step7
- Added archive https://web.archive.org/web/20110511091253/https://www.cambridgedna.com/genealogy-dna-ancient-migrations-slideshow.php?view=step7 to https://www.cambridgedna.com/genealogy-dna-ancient-migrations-slideshow.php?view=step7
- Added archive https://web.archive.org/web/20100703045010/http://nitro.biosci.arizona.edu/zdownload/papers/FSI-2008.PDF to http://nitro.biosci.arizona.edu/zdownload/papers/FSI-2008.PDF
- Added archive https://web.archive.org/web/20060221332211/http://anthro.palomar.edu/vary/vary_3.htm to http://anthro.palomar.edu/vary/vary_3.htm
- Added archive https://web.archive.org/web/20080405232839/https://www3.nationalgeographic.com/genographic/atlas.html to https://www3.nationalgeographic.com/genographic/atlas.html
When you have finished reviewing my changes, you may follow the instructions on the template below to fix any issues with the URLs.
This message was posted before February 2018. After February 2018, "External links modified" talk page sections are no longer generated or monitored by InternetArchiveBot. No special action is required regarding these talk page notices, other than regular verification using the archive tool instructions below. Editors have permission to delete these "External links modified" talk page sections if they want to de-clutter talk pages, but see the RfC before doing mass systematic removals. This message is updated dynamically through the template {{source check}} (last update: 5 June 2024).
- If you have discovered URLs which were erroneously considered dead by the bot, you can report them with this tool.
- If you found an error with any archives or the URLs themselves, you can fix them with this tool.
Cheers.—InternetArchiveBot (Report bug) 05:49, 9 January 2017 (UTC)
External links modified
[edit]Hello fellow Wikipedians,
I have just modified 6 external links on Genetic history of indigenous peoples of the Americas. Please take a moment to review my edit. If you have any questions, or need the bot to ignore the links, or the page altogether, please visit this simple FaQ for additional information. I made the following changes:
- Added archive https://web.archive.org/web/20131117011216/http://www.genebase.com/learning/article/16 to http://www.genebase.com/learning/article/16
- Added archive https://web.archive.org/web/20091104070218/http://www.genetree.com/education/q to http://www.genetree.com/education/q
- Added archive https://web.archive.org/web/20100617184224/http://usmex.ucsd.edu/assets/022/10143.pdf to http://usmex.ucsd.edu/assets/022/10143.pdf
- Added archive https://web.archive.org/web/20130505072339/http://www.andaman.org/BOOK/chapter54/text-Fuego/Genetics/text-FuegianGenetics.htm to http://www.andaman.org/BOOK/chapter54/text-Fuego/Genetics/text-FuegianGenetics.htm
- Added archive https://web.archive.org/web/20100226021520/http://www.metisnation.ca/who/index.html to http://www.metisnation.ca/who/index.html
- Corrected formatting/usage for http://anthro.palomar.edu/vary/vary_3.htm
When you have finished reviewing my changes, you may follow the instructions on the template below to fix any issues with the URLs.
This message was posted before February 2018. After February 2018, "External links modified" talk page sections are no longer generated or monitored by InternetArchiveBot. No special action is required regarding these talk page notices, other than regular verification using the archive tool instructions below. Editors have permission to delete these "External links modified" talk page sections if they want to de-clutter talk pages, but see the RfC before doing mass systematic removals. This message is updated dynamically through the template {{source check}} (last update: 5 June 2024).
- If you have discovered URLs which were erroneously considered dead by the bot, you can report them with this tool.
- If you found an error with any archives or the URLs themselves, you can fix them with this tool.
Cheers.—InternetArchiveBot (Report bug) 18:33, 12 October 2017 (UTC)
Major revision needed
[edit]The article, like most of our "genetic history" articles, needs a major revision based on literature published after 2010. In the early 2000s, "genetic history" studies mostly focussed on Y-DNA and mt-DNA. This wasn't because this is particularly relevant, but simply because the technology for autosomal studies had not been developed enough. Since c. 2010, the technology has been available, and results of unprecedented quality and confidence have been appearing since 2015 or so. All the "Y-DNA, mt-DNA" material is still valid, as far as it goes, but it is no longer of particular interest for genetic genealogy. Most of the material collected on this during the 2000s should be delegated to dedicated pages such as Y-DNA haplogroups in indigenous peoples of the Americas with only brief WP:SS summaries left in the main articles. --dab (𒁳) 10:38, 26 July 2018 (UTC)
Racial exogamy
[edit]The article states "Native Americans in the United States are more likely than any other racial group to practice racial exogamy ..." That is an extremely overreaching claim to make without a scientific study as a reference. In fact the statement has no reference at all. Mensch (talk) 00:32, 22 May 2019 (UTC)
- How about this one: "...Pew Research survey finds that biracial adults with a white and American Indian background comprise half of the country’s multiracial population—by far the country’s largest multiracial group..." [7] Rmhermen (talk) 07:34, 22 May 2019 (UTC)
Please update with: "Siberians Reveal Connections with First Americans and across Eurasia"
[edit]Could you please update the page to contain information on this study, included like so in 2020 in science:
Scientists report that genome-wide data of 19 Siberians of the Upper Paleolithic to Bronze Age of up to ca. 14,000 years ago show the most deeply divergent connection between Upper Paleolithic Siberians and the indigenous peoples of the Americas and that long-range human mobility across Eurasia during the Early Bronze Age as well as prolonged local admixture that lead to an ancestry that gave rise to all non-Arctic Native Americans.[1][2][3][4]
The news report has:
Past studies have indicated a connection between Siberian and American populations, but a 14,000-year-old individual analysed in this study is the oldest to carry the mixed ancestry present in Native Americans.
Furthermore, if you see a problem with the item's content at or notability for 2020 in science please edit it.
Thank you.
--Prototyperspective (talk) 14:13, 23 July 2020 (UTC)
References
- ^ "Oldest connection with Native Americans identified near Lake Baikal in Siberia". phys.org. Retrieved 13 June 2020.
- ^ "The oldest genetic link between Asians and Native Americans was found in Siberia". Science News. 20 May 2020. Retrieved 13 June 2020.
- ^ "Scientists discover oldest link between Native Americans, ancient Siberians". UPI. Retrieved 13 June 2020.
- ^ Yu, He; Spyrou, Maria A.; Karapetian, Marina; Shnaider, Svetlana; Radzevičiūtė, Rita; Nägele, Kathrin; Neumann, Gunnar U.; Penske, Sandra; Zech, Jana; Lucas, Mary; LeRoux, Petrus; Roberts, Patrick; Pavlenok, Galina; Buzhilova, Alexandra; Posth, Cosimo; Jeong, Choongwon; Krause, Johannes (11 June 2020). "Paleolithic to Bronze Age Siberians Reveal Connections with First Americans and across Eurasia" (PDF). Cell. 181 (6): 1232–1245.e20. doi:10.1016/j.cell.2020.04.037. ISSN 0092-8674. Retrieved 13 June 2020.
A Commons file used on this page or its Wikidata item has been nominated for deletion
[edit]The following Wikimedia Commons file used on this page or its Wikidata item has been nominated for deletion:
Participate in the deletion discussion at the nomination page. —Community Tech bot (talk) 19:47, 5 September 2020 (UTC)
Please update to include the European dna in founding populations
[edit]There are alot of errors in this post . Most of studies seem old. The topic needs to include the clear finding of European ancestry In founding popuations of Amerindians before Columbian contact meaning many were already mixed before with Asia and European dna. Where is the malta boys information. He was European and had a European phenotype. haplogroups are not races or phenotypes. had already written a post but it seems it has magically disappeared. I had several scientific articles added 107.190.109.82 (talk) 18:45, 4 September 2022 (UTC)
- Mal’ta boy wasn’t European. And is mentioned in this article. I gather you haven’t even read this article carefully. Doug Weller talk 19:03, 4 September 2022 (UTC)
South China
[edit]This editor repeatedly restores the following contribution:
Overall, the 'Ancestral Native Americans' formed from an 'Ancestral Native American' lineage which diverged from East Asian people around 36,000 years ago somewhere in Southern China, and subsequently migrated northwards into Siberia and encountered/interacted with a distinct Paleolithic Siberian population known as Ancient North Eurasians, closer related to modern Europeans, giving rise to both Indigenous peoples of Siberia and Native Americans.
However there does not appear to be anything in any study provided so far that says anything about an ancestral native American lineage arising in "Southern China". Nor does the sapiens.org link say that. It is strange that this editor apparently thinks that a web magazine is a more reliable source than a Cold Spring Harbor Laboratory study.
I would also point out the similarities of editing here with a banned IP, which also postulated a soufheast Asian origin for ancestral Native Americans that isn't found in the cited studies.[9][10] -- Hunan201p (talk) 10:41, 4 April 2023 (UTC)
- Sorry, the Southern China part is sourced by this paper[1]. Either including this cite, or remove the part about Southern China. The reference in question (above) is a review book rather a primary paper. My main concern is that the percentage and exact admixture patterns vary between different studies and models, therfore my initial inclusion of 42% ANE and 58% East Asian from the Wong paper is probably misleading (as other papers give different amounts and time periods). Btw. here is the book link:[11]. Thanks.94.131.108.230 (talk) 10:47, 4 April 2023 (UTC)
- There is good consensus that the East Asian contribution to the Native American and Paleo-Siberian ancestries stems from northern East Asia (probably the Amur area). The paper by X. Zhang et el. (2022) about the Red Deer Cave specimen (MZR = Mengzi Ren, as they call it) can be really terribly easily misunderstood. They just say that MZR is part of the wider E. Asian cluster that contributed to the Native American genepool, but not that MZR ancestry played a prominent part in it (i.e. more prominent than let's say Amur ancestry). –Austronesier (talk) 10:59, 4 April 2023 (UTC)
- Austronesier is 100% correct. There is nothing in Zhang, et al. (2022) that substantiates this claim:
'Ancestral Native Americans' formed from an 'Ancestral Native American' lineage which diverged from East Asian people around 36,000 years ago somewhere in Southern China
. - What they do say is that the Red Deer Cave people form one part of the amalgamation of Native American ancestry. The IP editor on the other hand speaks as if "Ancestral Native Americans" began with that one population. That is original research, and wrong. Even if Native Americans were direct descendants of Red Deer Cave, which they are not, the actual "Ancestral Native Americans" originate from the mixing of multiple populations, not from Red Deer Cave or ancient East Asians alone. And this mixing happened north of China. As Austronesier pointed out, it involved Ancient East Asians in Northeast Asia rather than southern China.
- The "northward migration and mixing with ANE" scenario is original research; these papers don't tell this tale.
- Further, regarding [12], their source cites Raghavan 2014 for the percentage of ANE-related ancestry in Paleo-Americans:
- Austronesier is 100% correct. There is nothing in Zhang, et al. (2022) that substantiates this claim:
- There is good consensus that the East Asian contribution to the Native American and Paleo-Siberian ancestries stems from northern East Asia (probably the Amur area). The paper by X. Zhang et el. (2022) about the Red Deer Cave specimen (MZR = Mengzi Ren, as they call it) can be really terribly easily misunderstood. They just say that MZR is part of the wider E. Asian cluster that contributed to the Native American genepool, but not that MZR ancestry played a prominent part in it (i.e. more prominent than let's say Amur ancestry). –Austronesier (talk) 10:59, 4 April 2023 (UTC)
Extended content
|
|---|
|
Our tree model grouped ANEs MA-1 and AG-2 with Western Siberians (Mansi and Nenets), suggesting a genetic link between ANEs and modern Western Siberians (Fig. 7). Furthermore, TreeMix estimated that 41% (95% CI: 36%–45%) of the ancestry of Andean Highlanders, the Native Americans, is attributable to the ANE lineage (Fig. 7; Supplemental Fig. S16). This is consistent with previous reports that Paleo-Americans trace ∼42% of their autosomal genome to an ancient Eurasian lineage related to MA-1 (Raghavan et al. 2014).
|
- ...so this is actually a secondary source for this figure. My main problem is that the IP has listed the ancestral components as "sister lineages" of East Asians and Europeans, when it is more accurate to describe them as ancient East Asians and ancient West Eurasians, or just ancient Eurasians. - Hunan201p (talk) 15:30, 4 April 2023 (UTC)
- The entire section "Autosomal DNA" probably needs a complete rewrite. I assume that all of this was assembled in good faith (and I'm not going to whine about socks (NB: atm!), since their contributions also look like good faith efforts, even when erroneous at times). But it has become all bloated and repetitive. Take e.g. the two paragraphs that cite content from Moreno-Mayar et al. (2018). Apart from being repetitive, they also contain the 36,000BCE date which tbh I can't find in the paper; maybe it's just me being dumb, so I'd be happy to see a quote in support of it from that paper.
- There are basically two models that describe the ancestral makeup of Ancestral Native Americans: in the fine-grained model by Sikora et at. (2019), Mao et al. (2021), the ANE ancestry component of Ancestral Native Americans is a sister lineage of MA-1 and AG3, i.e. admixed from the predominant West Eurasian source and a smaller East Eurasian (or East Asian) Tianyuan-related source, while the East Asian bulk of Ancestral Native American ancestry comes from a Devil's Cave-related north East Asian source. A more coarse model taps the ANE-like component directly from the West Eurasian source without the East Eurasian admixture observed in Yana, MA-1 and AG3 (Moreon-Mayar et al. 2018; Yu et al. 2020).
- Regadless of the model, calling this West Eurasian source a "sister lineage of Europeans" is just a matter of taste IMO. Some sources do that, some don't. I find it a bit anachronistic, since at the time when Ancestral Native American formed, much of Europe was still inhabited by populations that contributed little or nothing to the modern-day European gene pool. –Austronesier (talk) 16:39, 4 April 2023 (UTC)
- ...so this is actually a secondary source for this figure. My main problem is that the IP has listed the ancestral components as "sister lineages" of East Asians and Europeans, when it is more accurate to describe them as ancient East Asians and ancient West Eurasians, or just ancient Eurasians. - Hunan201p (talk) 15:30, 4 April 2023 (UTC)
- Hello, as I said at the talk page there, I have no problems with the part about Southern China to be removed. The other part is sourced by this 2022 book by Jennifer Raff:[2]
AROUND 36,000 years ago, a small group of people living in East Asia began to break off from the larger ancestral populations in the region. By about 25,000 years ago, the smaller group in East Asia itself split into two. One gave rise to a group referred to by geneticists as the ancient Paleo-Siberians, who stayed in Northeast Asia. The other became ancestral to Indigenous peoples in the Americas.
- I cited the summary article in Sapiens ([13]).
- Hello, as I said at the talk page there, I have no problems with the part about Southern China to be removed. The other part is sourced by this 2022 book by Jennifer Raff:[2]
- Another problem with the Wong paper may be that it is about Indigenous Andean Americans, rather than Indigenous Mexican groups. Other papers estimated different amounts of ANE ancestry for Native Americans. This (Moreno-Mayar et al. 2018)[3] for example estimate between 14% and 38% of Native American ancestry may originate from gene flow from the Mal'ta–Buret' (ANE) population, and say:
Using demographic modelling, we infer that the Ancient Beringian population and ancestors of other Native Americans descended from a single founding population that initially split from East Asians around 36 ± 1.5 ka, with gene flow persisting until around 25 ± 1.1 ka. Gene flow from ancient north Eurasians into all Native Americans took place 25–20 ka, with Ancient Beringians branching off around 22–18.1 ka.
- The book I cited, probably cites Moreno-Mayar et al. on that. Hope that helps.94.131.108.230 (talk) 17:44, 4 April 2023 (UTC)
- Oh my dumb indeed, I looked up in the wrong paper (Moreno-Mayar et al. 2018 in Science[14]) in my files LOL. The Nature paper plus Raff's book are a strong pair of sources. But in any case, we really need to consolidate the info. It's repeated three(!) times now. –Austronesier (talk) 18:30, 4 April 2023 (UTC)
- Another problem with the Wong paper may be that it is about Indigenous Andean Americans, rather than Indigenous Mexican groups. Other papers estimated different amounts of ANE ancestry for Native Americans. This (Moreno-Mayar et al. 2018)[3] for example estimate between 14% and 38% of Native American ancestry may originate from gene flow from the Mal'ta–Buret' (ANE) population, and say:
References
West Eurasian or ANE on Native American was removed.
[edit]R1b and mtDNA X are commonly found in west eurasia. It is found in Kennewick Man , his mtDNA is X
WHY IS THIS REMOVED?
A 2013 study in Nature reported that DNA found in the 24,000-year-old remains of a young boy from the archaeological Mal'ta-Buret' culture suggest that up to one-third of Indigenous Americans' ancestry can be traced back to western Eurasians, who may have "had a more north-easterly distribution 24,000 years ago than commonly thought"[1] "We estimate that 14 to 38 percent of Indigenous American ancestry may originate through gene flow from this ancient population," the authors wrote. Professor Kelly Graf said,
"Our findings are significant at two levels. First, it shows that Upper Paleolithic Siberians came from a cosmopolitan population of early modern humans that spread out of Africa to Europe and Central and South Asia. Second, [Paleo-Indigenous American] skeletons like Buhl Woman with phenotypic traits atypical of modern-day Indigenous Americans can be explained as having a direct historical connection to Upper Paleolithic Siberia.
Gemmaso (talk) 13:35, 16 April 2023 (UTC) Gemmaso (talk) 13:47, 16 April 2023 (UTC)
- @Gemmaso: it was removed because it is original research. It was placed in the "Haplogroup R1" sub-section, implying that R1 in Native Americans could be from ANE; when no source says that. Every source in the article that contemplates the origin of R1 in Native Americans says that it is most likely of post-1492 origin. See citations [55] and [56]. C and Q are the only founding paternal lineages in Native Americans.
- Haplogroup X2a in Native Americans is not of European origin and hasn't been found anywhere in Eurasia, but is most closely related to South Siberian X lineages, implying it was part od the same foubding population as A,B, C and D. This is explained in citation [72]. The fact that West Eurasians have other mutations of X doesn't make Native Americans related to them; West Eurasians belong to clades of X that are phylogenetically unrelated to Native American X lineages.
- Raff also point out in citation [72] that Kenneeick man had no extra dose of West Eurasian ancestry to go along with his haplogroup X.
- Unsurprisingly, there's never been an ANE specimen found with haplogroup X or R1. Most identified so far carried paternal haplogroup Q and the maternal haplogroup U. - Hunan201p (talk) 14:02, 16 April 2023 (UTC)
Extended content
|
|---|
|
Malhi et al. 2008:
All individuals that did not belong to haplogroup Q and C were excluded from the Haplotype data set because these haplotypes are likely the result of non-native admixture (Tarazona-Santos and Santos, 2002; Zegura et al., 2004; Bolnick et al, 2006)...The frequency of haplogroup C is highest in Northwestern North America and the frequency of haplogroup R, the presence of which is attributed to European admixture, reaches its maximum in Northeastern North America." Raff 2022, p. 59-60:"Y chromosome founder haplogroups in Native Americans include Q-M3 (and its sub-haplogroups, Q-CTS1780), and C3-MPB373 (potentially C- P39-Z30536). Other haplogroups found [sic] Native American populations, like R1b, were likely the result of post-European contact admixture (44)." |
- Hunan201p (talk) 14:09, 16 April 2023 (UTC)
So you mean all these years it was original research? Since 2013 many sources had stated 1/3 of Native American genes come from west eurasian people's, for example.- https://www.nationalgeographic.com/science/article/131120-science-native-american-people-migration-siberia-genetics
- " Nearly one-third of Native American genes come from west Eurasian peoples with ties to the Middle East and Europe"
- Every source in the article that contemplates the origin of R1 in Native Americans says that it is most likely of post-1492 origin
- According to what source and how is that possible. If R1b in Native American came post -1492 origin, how is it that nearly 92-98% of Native Americans west eurasian lineage are R1b, with nearly no other west eurasian y-dna haplogroups. How can R1b be so high?
- Haplogroup X2a even if it was closely related with South Siberian X lineages, ancient South Siberians were R1a related with ancient Indo-Europeans. It is impossible to associate mtDNA X lineages with East Eurasians as the original original X were clearly at west Eurasians or at least a mixed with East-West(just in case). Ancient South Siberians haplogroup y-dna, majority were R1a.
- " A more ancient source is presumably the South Siberian and/or Central Asian haplotypes brought to the Hindustan during the westward migrations of R1a bearers between 20,000 and 10,000 ybp. "
- (Source:Anatole A. Klyosov · 2018)
- "Finally, our data indicate that at the Bronze and Iron Age timeframe, south Siberians were blue (or green)-eyed, fair-skinned and light-haired people"
(Source: Ancient DNA:history of south Siberian"Gemmaso (talk) 14:24, 16 April 2023 (UTC)- @Gemmaso: in response to your points:
- There is no evidence that haplogroup X was "originally West Eurasian".
- Klyosov is not considered a reliable source on Wikipedia, and is all but deprecated, for various reasons.
- The reason why middle-late Bronze Age inhabitants of South Siberia had R1a/blue eyes/blond hair combo, is primarily because they were recent migrants from the West Eurasian steppe. They were Western Steppe Herders, not ANE, whose complete pigmentation variation is unknown. - Hunan201p (talk) 14:34, 16 April 2023 (UTC)
- @Gemmaso: in response to your points:
What do you mean there is no evidence? X is found 2% in Europe and is also found in moderate-high frequencies North Africa, Middle east, Caucasus, Horn of Africa. X is found , 14.3%, among the natives of Bahariya Oasis (Western Desert, Egypt) X2 alone is found 11% in Druze people. Druze in the Assyrian community in Israel 27% others also found in Georgia (8%), Orkney (Scotland) (7%), It's been found in ancient Egyptian mummies and ancient Assyrians?- In ancient and Iberia Chalcolithic (X2b, La Chabola de la Hechicera, 1/3 or 33%; X2b, El Sotillo, 1/3 or 33%; X2b, El Mirador Cave, 1/12 or ~8%) cultures
- EVEN IF X in Native Americans had arisen from early-Paleo Indian, it is clearly to anyone that Kennewick Man who is mtDNA X did not look Native American, anthropologist said he looked European. I have no objection if you want to claim mtDNA X in Native American is not of European origin but it should at the very least stated that mtDNA X is not a haplogroup that is found in East Asian/East Eurasian, is non-existant to them. It's found only in west eurasian populations in Europe and Asia or East-West populations(admixed).
- To reply your exented content
Extended content
|
|---|
|
Raghavan et al. 2014
|
Your 2002, 2004, 2006 studies are outdated studies that did not know about the connection of Malta boy and Native Americans. Your 2022 also only say they were likely post-1492 contribution but is not definite. There is not reason to remove the 2014 study that suggests a possibility that R1b in Native American is one of the founding lineages. Gemmaso (talk) 14:56, 16 April 2023 (UTC)- The fact that haplogroup X is found in Europe does not make it "West Eurasian". The haplogroup is thought to have arisen in Africa and Asia, but whether it originated in the ancestors of West Eurasians or East Eurasians is unknown. As you know, there were genetically "un-West Eurasian" people in West Eurasia in those days, such as Oase. But what is truly important here is that X2a is not West Eurasian. It is East Eurasian and only found in East Eurasians (Native Americans), per Raff (2015) it evolved in East Eurasia.
- Your source regarding Raghavan (2014) does not say or even imply that Mal'ta boy could explain R1 in Native Americans. As your own quote says, it speaks only of autosomal evidence. The 2000s studies are not outdated, because the existence of Mal'ta boy isn't needed to show that Native American R1 is from post-1492 admixture. That's because MRCA dating and European-specific phylogeny easily demonstrates that it is. - Hunan201p (talk) 15:16, 16 April 2023 (UTC)
I won't debate anymore if X is west Eurasian, it certainly not distributed in East Eurasians. The fact the article still must state " haplogroup X is the only haplogroup that is not connected to East Asia, it found exclusively in Europe, Middle East, Caucasus, Africa. At least stress the importance. Here is a study supporting X2 is West Eurasian https://www.nature.com/articles/jhg201164 " West Eurasian mtDNA haplogroups found in gene pools of South Siberians (for example, N1e, I4, J1b2, N1a, U4 and X2e)"- Malt'a boy is R1b. And if that doesn't have to do with Native American West Eurasian admixture. Than what on earth does? Native Americans clearly have different nose types and facial structure, appearance from East Asians.
- https://greekreporter.com/2022/11/19/dna-native-americans-western-eurasia/
- "However, what is so groundbreaking about the DNA study is that Native Americans appear to have been a mixed group, having ancestors directly from Western Eurasia as well as the forebears of those who are now living in Eastern Asia. “The meeting of those two groups is what formed Native Americans as we know them,” Willerslev declared."
- “Although we know that North Americans are related to East Asians, it’s striking that no contemporary East Asian populations really resemble Native Americans,” Willerslev explained.
- “It’s not like you can say that they are really closely related to Japanese, Chinese, or Koreans, so there seems to be something missing,” Willerslev said. “But this result makes a lot of sense regarding why they don’t fit so well genetically with contemporary East Asians —because one-third of their genome is derived from another population.” "
Gemmaso (talk) 15:41, 16 April 2023 (UTC)
- Stressing that X isn't East Eurasian would be futile since the secondary source already cited in the article (Raff 2015) explicitly lines out that
"Although X2a's ancestor (X2) is thought to have evolved in the Near East (part of “West Eurasia”), characterizing X2a as a “West Eurasian” haplogroup is inaccurate because X2a is found only in people with indigenous North American ancestry. Associating X2a with “West Eurasia” is like saying “Solutreans evolved in Africa”: each statement refers to a location where the ancestral population is thought to have lived long ago, but that location is not relevant to the question under consideration. In particular, because X2 dispersed from the Near East and became widely distributed throughout the world, its descendent lineage X2a need not have evolved in the same place."
Native Americans belong to X2a not X2e. - Mal'ta boy was not R1b, he belonged to R* which is not confirmed to exist in any living person.
- Your source doesn't say that ANE can explain R1 in Native Americans. The phylogenic evidence clearly shows that R1 in Native Americans is from post-1492 colonization.
- Per Segura, et al., page 169:
- Stressing that X isn't East Eurasian would be futile since the secondary source already cited in the article (Raff 2015) explicitly lines out that
Figure 5 gives the network for haplogroup R-P25 in Europe, Asia, and the Americas. The large central node represents 12 individuals (4 Sioux, 2 Mixtec, 1 Cheyenne, 1 Wayu, 1 Greek, 1 Italian, 1 Russian, and 1 Britain) deriving from four Native American and four European populations and exhibiting haplotype (14–12–13–29–24–11–13–13–12–12), which is identical to the R-P25 modal haplotype forboth Native Americans and Europeans. In contrast, this modal haplotype differs from the Asian modal haplotype at two positions (DYS390 ¼ 23; DYS393 ¼ 12). Extensive sharing of haplotypes between Native Americans and Europeans is evident throughout the network.
- ...and there is also the fact that R1b frequency in Native Americans shows a clear gradient from East-to-West in North America, which was mentioned in Malhi, et al (2008). - Hunan201p (talk) 16:07, 16 April 2023 (UTC)
How is it futile when X is absent in East Eurasian population( pure ones anyone)?- Like I said: It doesn't matter if X2a is considered west eurasian or not. We should still state mtDNA X or X2 is not associated with East Asia or East Eurasian. It is completely absent in their world.
- YES, X2a is found only in people with indigenous North American ancestry but these people are native Americans ( people who have 1/3 West Eurasian or 38%), there is no genetic study that claims X2a came from pure East Eurasian population from East Asia.
- So even if X in Native American is not regarded as West Eurasian origin because some conspiracy theories suggested it was related to Solutreans(which is now debunked), it still doesn't change the fact that is distributed all over west eurasia populations even in Africa's west eurasians. According to the nature.com study, haplogroup mtDNA X in Native American and South Siberians are West Eurasians and so this explains partially why Native Americans have west eurasian admixtures. Anyway Raff 2015 own opinion shouldn't exclude others aswell.
- Now as for R1b. Native Americans most common R1b is M173 and the 2002 study does show a Siberian connection".
- READ THIS
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC384887/
- The Y chromosomes of 549 individuals from Siberia and the Americas were analyzed for 12 biallelic markers, which defined 15 haplogroups.
- The second major group of Native American Y chromosomes, haplogroup M45, accounted for about one-quarter of male lineages. M45 was subdivided by the biallelic marker M173 and by the four microsatellite loci alleles into two major subdivisions: M45a, which is found throughout the Americas, and M45b, which incorporates the M173 variant and is concentrated in North and Central America. In Siberia, M45a haplotypes, including the direct ancestor of haplogroup M3, are concentrated in Middle Siberia, whereas M45b haplotypes are found in the Lower Amur River and Sea of Okhotsk regions of eastern Siberia. Among the remaining 5% of Native American Y chromosomes is haplogroup RPS4Y-T, found in North America. In Siberia, this haplogroup, along with haplogroup M45b, is concentrated in the Lower Amur River/Sea of Okhotsk region. These data suggest that Native American male lineages were derived from two major Siberian migrations. The first migration originated in southern Middle Siberia with the founding haplotype M45a (10-11-11-10). In Beringia, this gave rise to the predominant Native American lineage, M3 (10-11-11-10), which crossed into the New World. A later migration came from the Lower Amur/Sea of Okhkotsk region, bringing haplogroup RPS4Y-T and subhaplogroup M45b, with its associated M173 variant. This migration event contribute
- The M45b/M173 subhaplogroup connects eastern Siberians with the North and Central American Na-Dene and surrounding Amerinds (fig. 3).
- Anyway there is still tons of study that supports a pre-columbian origin.
Gemmaso (talk)16:08, 16 April 2023 (UTC)Something you should know about R-P25 (which is from your 2004 study)- " before 2005, R1b was synonymous with R-P25, which was later reclassified as R1b1; in 2016, R-P25 was removed completely as a defining SNP, due to a significant rate of back-mutation. (Below is the basic outline of R1b according to the ISOGG Tree as it stood on January 30, 2017.) " Gemmaso (talk) 17:12, 16 April 2023 (UTC)
- This should debunk any claims of Native Americans R-M173 being likely the result of post-European contact admixture
- http://en.wiki.x.io/wiki/Haplogroup_R-M269
- Haplogroup R-M269 which is the predominant (90%) haplogroup R1b of west Europeans. You can see the table.
- In Spain it reach 87.1%, in England 62%, Portugal 46.2%, Germany 32.2%. This R-M269 is also found over 46% of White European Americans, and 14% of African Americans. The Highest are in the Wales at 92.3% but there literally no contact between wales and any Amerindian.
- The other one is R-U16 which makes up 20% in England, 19% Germany, 15% White European Americans, 2.5% African Americans
In a sample of 33 it shows Central/South America it's 0% R-M269 and 0% R-116. This should be impossible given that West Europeans were all over the place in Central and South America. I could be wrong but so far I can't find anything that suggest it.Gemmaso (talk) 17:48, 16 April 2023 (UTC)
- B-Class Molecular Biology articles
- Unknown-importance Molecular Biology articles
- B-Class MCB articles
- Mid-importance MCB articles
- WikiProject Molecular and Cellular Biology articles
- All WikiProject Molecular Biology pages
- B-Class Anthropology articles
- Mid-importance Anthropology articles
- B-Class Ethnic groups articles
- High-importance Ethnic groups articles
- WikiProject Ethnic groups articles
- B-Class Indigenous peoples of North America articles
- High-importance Indigenous peoples of North America articles
- WikiProject Indigenous peoples of North America articles
- B-Class Arctic articles
- Mid-importance Arctic articles
- WikiProject Arctic articles
- B-Class Alaska articles
- High-importance Alaska articles
- WikiProject Alaska articles
- B-Class Canada-related articles
- Mid-importance Canada-related articles
- B-Class History of Canada articles
- Mid-importance History of Canada articles
- All WikiProject Canada pages
- B-Class Greenland articles
- Mid-importance Greenland articles
- WikiProject Greenland articles
- B-Class South America articles
- Mid-importance South America articles
- WikiProject South America articles
- B-Class Indigenous peoples of the Americas articles
- Unknown-importance Indigenous peoples of the Americas articles
- Indigenous peoples of the Americas articles
- B-Class Caribbean articles
- Mid-importance Caribbean articles
- WikiProject Caribbean articles
- B-Class United States articles
- Mid-importance United States articles
- B-Class United States articles of Mid-importance
- B-Class United States History articles
- Unknown-importance United States History articles
- WikiProject United States History articles
- WikiProject United States articles













