Draft:Virome analysis
 | Review waiting, please be patient.
This may take 2 months or more, since drafts are reviewed in no specific order. There are 2,337 pending submissions waiting for review.
Where to get help
How to improve a draft
You can also browse Wikipedia:Featured articles and Wikipedia:Good articles to find examples of Wikipedia's best writing on topics similar to your proposed article. Improving your odds of a speedy review To improve your odds of a faster review, tag your draft with relevant WikiProject tags using the button below. This will let reviewers know a new draft has been submitted in their area of interest. For instance, if you wrote about a female astronomer, you would want to add the Biography, Astronomy, and Women scientists tags. Editor resources
Reviewer tools
|
Virome analysis refers to the study of all viral material in an organism or ecosystem. The virome can be described as the collection of total viruses found in an organism or ecosystem.[1] Viromes are incredibly diverse, varying between and within sample types, and are often poorly characterized.[2] Virome diversity is influenced by factors such as lifestyle, age, diet, and genetic diversity to name a few.[3] Studying viromes across sample types has demonstrated unique interactions between host-virus and virus-microbiome.[4] These interactions contribute to the overall health and disease of an individual either through infecting the host, or indirectly through modulating microbial communities (bacteriophages).[4]
Background
[edit]History
[edit]
The first virome analysis was performed in 2002 investigating virus composition of seawater samples collected off the coast of California.[1] More than 65% of the viral sequences had not been seen before, highlighting the viral diversity of environmental viromes.[1] Between 2003 and 2006, similar metagenomic experiments in human fecal samples exploring the human virome yielded comparable rates of viral diversity including an abundance of viral 'dark matter'.[6][7] These early studies relied on Sanger sequencing and were limited in both throughput and sequencing depth but supported the emergence of virome analysis.[1][6][7] The development of next-generation sequencing (NGS) greatly expanded virome analysis capabilities and knowledge on virome diversity.[8] Metagenomic shotgun sequencing is often used in virome studies as an unbiased approach for sequencing the total viral communities of the sample.[3] This sequencing approach produces shorter reads (~100 - 300 bp) but can generate millions of reads drastically improving the sequencing depth and coverage. These metagenomic studies allow for viral discovery, classification, and exploration of host-virus interactions, but are greatly limited by the computational analysis.[9][10]
Traditional virome analysis
[edit]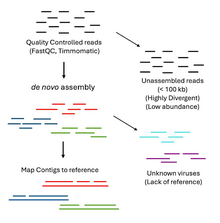
The output of virome metagenomic studies using shotgun sequencing is hundreds of thousands or even millions of short reads (~100 - 300 bp). These reads undergo quality control checkpoints using tools to assess sequence read quality, read trimming and host depletion to prepare the viral sequences for assembly and alignment.[2] Reference-guided de novo assembly is the most popular method for genome assembly in virome analysis.[11] Sequencing reads are assembled into overlapping subsequences of a fixed length k (k-mers) known as contigs.[11][12] Contigs are aligned to reference databases for sequence similarity to assign viral taxonomy of the sample.[2] This method, however, requires prior knowledge of viral taxonomy and is greatly impacted by the lack of robust references available.[13] Current databases tend to be biased towards clinically relevant and cultivable viruses, notably reducing the analysis power.[13] As a result, it is believed that our understanding of virus classification and taxonomy greatly underestimates the virome's true diversity.[13]
Another limitation is the ability of the assembly tools to assemble low coverage, low abundance viruses.[11] Low abundance viruses may end up fragmented if sequencing depth is insufficient.[11] Tools can adjust for shorter k-mer lengths to include fragmented viral reads but this can introduce issues with contig ambiguity.[11] This limitation leads to considerable proportions of uncharacterized viral sequencing reads or 'viral dark matter'. New analysis software that harnesses machine learning have emerged to improve the deficiencies of reference database similarity approaches.[13]
Deep learning in virome classification
[edit]Deep learning has demonstrated advantages in many other applications within the genomics field, often surpassing traditional, state-of-the-art computational methods in terms of predictive performance, especially when trained with sufficient data.[14] Deep learning supports multitask learning, which is an approach where the model shares knowledge across a primary task and one or more secondary tasks, improving the versatility of tools.[15] Moreover, with multi-view learning, which facilitates the integration of multiple data types–such as sequence data, DNA methylation, gene expression, and more–can produce more accurate and robust predictions.[14]
Virome classification and analysis present a unique challenge due to the rapid evolution of viral genomes, which often leads to high sequence divergence within a species.[16] Deep learning models attempt to address this challenge and can recognize complex patterns in viral sequence fragments while handling high-dimensional data.[17]
Viral identification
[edit]Traditional database-based tools like BLAST rely on reference data and can struggle with highly divergent viruses with no known homologs across previously identified in existing genomes[18] – these sequences are generally classified as "unknown",[18] providing little information to the user. Similarly, other sequence alignment-based methods, such as Kraken[19] and Metavir,[20] also face limitations due to biases in databases. Current virus genome databases are heavily skewed towards viruses that infect hosts that are cultivable in the lab.[21] The lack of sufficient data available can negatively impact viral identification. For example, one study estimates that only 15% of viruses in the human gut have similarity to known viruses in databases,[21] limiting the extent of expected matches.
Several tools use traditional machine-learning approaches for viral identification. For example, HMMER3 uses profile Hidden Markov Models (pHMMs) based on reference databases of viral protein families to characterize unknown viruses.[22] However, this method is still constrained by the scarcity of characterized viral proteins in viral databases and can struggle with highly divergent viral sequences.[18] Deep learning provides a more flexible alternative, as models do not have to rely solely on predefined reference databases but instead, learn to recognize viral genomic signatures from the training data.[18]
Virome-host interaction analysis
[edit]Another important application of deep learning is virome-host interaction analysis. Currently, no high-throughput experimental methods can definitively assign a host to uncultivated viruses.[23] Alignment-based approaches struggle due to the scarcity of robust data in reference databases and high viral sequence divergence.[23] On the other hand, alignment-free methods– using features such as k-mer composition analysis, codon usage, and GC content, to measure similarity between viral and host sequences to other viruses with a known host, provide a viable alternative.[23] Since genomic features are embedded in viral genomes, deep learning models could learn these features automatically to drive predictions.[23] For example, evoMIL, which predicts virus-host association at the species level, accepts the viral sequence as a sole input.[24]
Viral resistance and mutation detection
[edit]Deep learning models can also be used to characterize drug resistance in viruses through the identification of drug resistance mutations.[25] Here models can make predictions and identify novel patterns in the input data, rather than relying on known drug resistance mutation.[25] Geometric deep learning, which incorporates physical knowledge into neural architectures,[26] could increase model prediction performance here, increasing the depth of learned patterns by incorporating 3-Dimensional molecular structure in drug interaction.[27]
Deep learning models used
[edit]Deep learning models applied in virome classification include, but are not limited to Convolutional Neural Networks (CNNs), Recurrent Neural Networks (RNNs), and Large Language Models (LLMs).
Convolutional neural networks (CNNs)
[edit]CNNs are widely used for sequence classification, processing inputs as multidimensional arrays, treating nucleotide sequences as unstructured data.[18] Each convolutional layer applies learnable filters that scan through the input sequence, extracting key features from local patterns.[18] Multiple filters are applied simultaneously, generating a set of feature maps.[18] Examples of CNN-based virome classification tools include DeepVirFinder[21] and ViraMiner[18] which both use a combination of CNNs and dense neural networks. DeepVirFinder learns viral genomic signatures and builds a predictive model based on these features.[21] It processes DNA sequences by encoding them, passing them through convolutional layers, applying max pooling and a fully connected layer, and ultimately outputting a probability score between 0 and 1 for binary classification.[21] ViraMiner uses a similar architecture but uses the average operator instead of the maximum operator to maintain more information about the frequency of patterns.[18]
Recurrent neural networks (RNNs)
[edit]Long Short-Term Memory (LSTM) architecture, a type of RNN, has been highly efficient for classification tasks despite being originally developed for generative tasks.[28] This has allowed the application of LSTMs in virome classification tasks.[28] LSTMs are effective for sequence-based tasks because they maintain long-term memory, allowing them to recognize long-range dependencies in the data when making predictions.[29] An example of an LSTM-based tool is ViroNIA, which predicts hepatitis C virus (HCV) sequences.[28] ViroNIA processes one-hot encoded viral sequences that are padded to a fixed length and then analyzed hierarchically with two LSTM layers.[28] Another model, Seeker, uses LSTM architecture to identify bacteriophages.[30]
Large language models (LLMs)
[edit]LLMs are a type of language model that uses neural networks trained on large quantities of unlabeled text data.[31] LLMs have been shown to have a variety of applications in genomics, including within the field of viromic analysis.[14] For example, a viral language model, ViraLM,[32] uses an adapted version of the genome foundation model, DNABERT-2,[33] for more efficient and accurate viral classification.
Future
[edit]Incorporating a multiomics approach into virome analysis could provide a more comprehensive understanding of the biology. Transcriptomics can assist in determining gene expression between genetically different viral strains leading to fitness within the virome, and virus-host interactions.[34] Analyzing viral transcripts can also help characterize viral infections and distinguish between latent or active infections.[34] Proteomics studies can confirm findings from transcriptomic studies and can determine biomarkers as diagnostic and therapeutic targets.[35] Metabolomics can provide valuable information on the biochemical changes due to the composition of viruses.[36] Metabolites produced by the host in response to viral infections can be used as biomarkers to help with predicting the virome diversity.[36] Virome analysis with the inclusion of multiomics can lead to improved personalized medicine through a more comprehensive understanding of the virome's role in a host.[36]
Another study direction is population wide virome surveillance to understand viral outbreaks. This can be achieved through using environmental matrices such as wastewater as a proxy to determine emerging viruses or circulation of high pathogenic strains.[37] Zoonotic spillover events could be predicted or detected through monitoring high-risk host reservoirs such as rodents, livestock or birds.[38] Surveillance of viruses is becoming increasingly important for outbreak prevention and investigation.
References
[edit]- ^ a b c d Breitbart, Mya; Salamon, Peter; Andresen, Bjarne; Mahaffy, Joseph M.; Segall, Anca M.; Mead, David; Azam, Farooq; Rohwer, Forest (2002-10-16). "Genomic analysis of uncultured marine viral communities". Proceedings of the National Academy of Sciences. 99 (22): 14250–14255. Bibcode:2002PNAS...9914250B. doi:10.1073/pnas.202488399. ISSN 0027-8424. PMC 137870. PMID 12384570.
- ^ a b c Wommack, K. Eric; Bhavsar, Jaysheel; Polson, Shawn W.; Chen, Jing; Dumas, Michael; Srinivasiah, Sharath; Furman, Megan; Jamindar, Sanchita; Nasko, Daniel J. (2012-07-27). "VIROME: a standard operating procedure for analysis of viral metagenome sequences". Standards in Genomic Sciences. 6 (3): 427–439. Bibcode:2012SGenS...6..421W. doi:10.4056/sigs.2945050. ISSN 1944-3277. PMC 3558967. PMID 23407591.
- ^ a b Liang, Guanxiang; Bushman, Frederic D. (2021-03-30). "The human virome: assembly, composition and host interactions". Nature Reviews Microbiology. 19 (8): 514–527. doi:10.1038/s41579-021-00536-5. ISSN 1740-1526. PMC 8008777. PMID 33785903.
- ^ a b Handley, Scott A. (2016-04-01). "The virome: a missing component of biological interaction networks in health and disease". Genome Medicine. 8 (1). doi:10.1186/s13073-016-0287-y. ISSN 1756-994X. PMID 27037032.
- ^ "Scientific Image and Illustration Software | BioRender". www.biorender.com. Retrieved 2025-02-21.
- ^ a b Breitbart, Mya; Hewson, Ian; Felts, Ben; Mahaffy, Joseph M.; Nulton, James; Salamon, Peter; Rohwer, Forest (2003-10-15). "Metagenomic Analyses of an Uncultured Viral Community from Human Feces". Journal of Bacteriology. 185 (20): 6220–6223. doi:10.1128/jb.185.20.6220-6223.2003. ISSN 0021-9193. PMC 225035. PMID 14526037.
- ^ a b Zhang, Tao; Breitbart, Mya; Lee, Wah Heng; Run, Jin-Quan; Wei, Chia Lin; Soh, Shirlena Wee Ling; Hibberd, Martin L; Liu, Edison T; Rohwer, Forest; Ruan, Yijun (2005-12-20). "RNA Viral Community in Human Feces: Prevalence of Plant Pathogenic Viruses". PLOS Biology. 4 (1): e3. doi:10.1371/journal.pbio.0040003. ISSN 1545-7885. PMID 16336043.
- ^ Dolja, Valerian V.; Koonin, Eugene V. (January 2018). "Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer". Virus Research. 244: 36–52. doi:10.1016/j.virusres.2017.10.020. ISSN 0168-1702. PMC 5801114. PMID 29103997.
- ^ Tyson, Gene W.; Chapman, Jarrod; Hugenholtz, Philip; Allen, Eric E.; Ram, Rachna J.; Richardson, Paul M.; Solovyev, Victor V.; Rubin, Edward M.; Rokhsar, Daniel S.; Banfield, Jillian F. (2004-02-01). "Community structure and metabolism through reconstruction of microbial genomes from the environment". Nature. 428 (6978): 37–43. Bibcode:2004Natur.428...37T. doi:10.1038/nature02340. ISSN 0028-0836. PMID 14961025.
- ^ Segata, Nicola; Boernigen, Daniela; Tickle, Timothy L; Morgan, Xochitl C; Garrett, Wendy S; Huttenhower, Curtis (January 2013). "Computational meta'omics for microbial community studies". Molecular Systems Biology. 9 (1). doi:10.1038/msb.2013.22. ISSN 1744-4292. PMID 23670539.
- ^ a b c d e Quince, Christopher; Walker, Alan W; Simpson, Jared T; Loman, Nicholas J; Segata, Nicola (September 2017). "Shotgun metagenomics, from sampling to analysis". Nature Biotechnology. 35 (9): 833–844. doi:10.1038/nbt.3935. ISSN 1087-0156. PMID 28898207.
- ^ Bankevich, Anton; Nurk, Sergey; Antipov, Dmitry; Gurevich, Alexey A.; Dvorkin, Mikhail; Kulikov, Alexander S.; Lesin, Valery M.; Nikolenko, Sergey I.; Pham, Son; Prjibelski, Andrey D.; Pyshkin, Alexey V.; Sirotkin, Alexander V.; Vyahhi, Nikolay; Tesler, Glenn; Alekseyev, Max A. (May 2012). "SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing". Journal of Computational Biology. 19 (5): 455–477. doi:10.1089/cmb.2012.0021. ISSN 1066-5277. PMC 3342519. PMID 22506599.
- ^ a b c d Ren, Jie; Ahlgren, Nathan A.; Lu, Yang Young; Fuhrman, Jed A.; Sun, Fengzhu (2017-07-06). "VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data". Microbiome. 5 (1). doi:10.1186/s40168-017-0283-5. ISSN 2049-2618.
- ^ a b c Yue, Tianwei; Wang, Yuanxin; Zhang, Longxiang; Gu, Chunming; Xue, Haoru; Wang, Wenping; Lyu, Qi; Dun, Yujie (2018), Deep Learning for Genomics: A Concise Overview, arXiv:1802.00810, retrieved 2025-02-21
- ^ Seltzer, Michael L.; Droppo, Jasha (May 2013). "Multi-task learning in deep neural networks for improved phoneme recognition". 2013 IEEE International Conference on Acoustics, Speech and Signal Processing. pp. 6965–6969. doi:10.1109/ICASSP.2013.6639012. ISBN 978-1-4799-0356-6.
- ^ Elbasir, Abdurrahman; Ye, Ying; Schäffer, Daniel E.; Hao, Xue; Wickramasinghe, Jayamanna; Tsingas, Konstantinos; Lieberman, Paul M.; Long, Qi; Morris, Quaid; Zhang, Rugang; Schäffer, Alejandro A.; Auslander, Noam (2023-02-11). "A deep learning approach reveals unexplored landscape of viral expression in cancer". Nature Communications. 14 (1): 785. Bibcode:2023NatCo..14..785E. doi:10.1038/s41467-023-36336-z. ISSN 2041-1723. PMID 36774364.
- ^ Sukhorukov, Grigorii; Khalili, Maryam; Gascuel, Olivier; Candresse, Thierry; Marais-Colombel, Armelle; Nikolski, Macha (2022-05-13). "VirHunter: A Deep Learning-Based Method for Detection of Novel RNA Viruses in Plant Sequencing Data". Frontiers in Bioinformatics. 2. doi:10.3389/fbinf.2022.867111. ISSN 2673-7647. PMC 9580956. PMID 36304258.
- ^ a b c d e f g h i Tampuu, Ardi; Bzhalava, Zurab; Dillner, Joakim; Vicente, Raul (2019-04-08). "ViraMiner: Deep Learning on Raw DNA Sequences for Identifying Viral Genomes in Human Samples". doi.org. doi:10.1101/602656. Retrieved 2025-02-21.
- ^ Wood, Derrick E.; Lu, Jennifer; Langmead, Ben (2019-09-07). "Improved metagenomic analysis with Kraken 2". doi.org. doi:10.1101/762302. Retrieved 2025-02-21.
- ^ Roux, Simon; Tournayre, Jeremy; Mahul, Antoine; Debroas, Didier; Enault, François (2014-03-19). "Metavir 2: new tools for viral metagenome comparison and assembled virome analysis". BMC Bioinformatics. 15 (1): 76. doi:10.1186/1471-2105-15-76. ISSN 1471-2105. PMC 4002922. PMID 24646187.
- ^ a b c d e Ren, Jie; Song, Kai; Deng, Chao; Ahlgren, Nathan A.; Fuhrman, Jed A.; Li, Yi; Xie, Xiaohui; Poplin, Ryan; Sun, Fengzhu (March 2020). "Identifying viruses from metagenomic data using deep learning". Quantitative Biology. 8 (1): 64–77. doi:10.1007/s40484-019-0187-4. ISSN 2095-4689. PMID 34084563.
- ^ Mistry, Jaina; Finn, Robert D.; Eddy, Sean R.; Bateman, Alex; Punta, Marco (2013-04-17). "Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions". Nucleic Acids Research. 41 (12): e121. doi:10.1093/nar/gkt263. ISSN 1362-4962.
- ^ a b c d Yan, Binghao; Nam, Yunbi; Li, Lingyao; Deek, Rebecca A.; Li, Hongzhe; Ma, Siyuan (2025-01-07). "Recent advances in deep learning and language models for studying the microbiome". Frontiers in Genetics. 15. doi:10.3389/fgene.2024.1494474. ISSN 1664-8021. PMC 11747409. PMID 39840283.
- ^ Liu, Dan; Young, Francesca; Robertson, David L; Yuan, Ke (2023-04-08). "Prediction of virus-host associations using protein language models and multiple instance learning". doi.org. doi:10.1101/2023.04.07.536023. Retrieved 2025-02-21.
- ^ a b Steiner, Margaret C.; Gibson, Keylie M.; Crandall, Keith A. (2020-05-19). "Drug Resistance Prediction Using Deep Learning Techniques on HIV-1 Sequence Data". Viruses. 12 (5): 560. doi:10.3390/v12050560. ISSN 1999-4915. PMID 32438586.
- ^ Bronstein, Michael M.; Bruna, Joan; Cohen, Taco; Veličković, Petar (2021), Geometric Deep Learning: Grids, Groups, Graphs, Geodesics, and Gauges, arXiv:2104.13478, retrieved 2025-02-21
- ^ Das, Bihter; Kutsal, Mucahit; Das, Resul (October 2022). "A geometric deep learning model for display and prediction of potential drug-virus interactions against SARS-CoV-2". Chemometrics and Intelligent Laboratory Systems. 229: 104640. doi:10.1016/j.chemolab.2022.104640. ISSN 0169-7439. PMC 9400382. PMID 36042844.
- ^ a b c d Ahmed, Hania; Mumtaz, Zilwa; Saqib, Sharmeen; Zubair Yousaf, Muhammad (March 2025). "ViroNia: LSTM based proteomics model for precise prediction of HCV". Computers in Biology and Medicine. 186: 109573. doi:10.1016/j.compbiomed.2024.109573. ISSN 0010-4825. PMID 39733555.
- ^ Van Houdt, Greg; Mosquera, Carlos; Nápoles, Gonzalo (2020-05-13). "A review on the long short-term memory model". Artificial Intelligence Review. 53 (8): 5929–5955. doi:10.1007/s10462-020-09838-1. ISSN 0269-2821.
- ^ Auslander, Noam; Gussow, Ayal B; Benler, Sean; Wolf, Yuri I; Koonin, Eugene V (2020-10-12). "Seeker: alignment-free identification of bacteriophage genomes by deep learning". Nucleic Acids Research. 48 (21): e121. doi:10.1093/nar/gkaa856. ISSN 0305-1048. PMID 33045744.
- ^ Raiaan, Mohaimenul Azam Khan; Mukta, Md. Saddam Hossain; Fatema, Kaniz; Fahad, Nur Mohammad; Sakib, Sadman; Mim, Most Marufatul Jannat; Ahmad, Jubaer; Ali, Mohammed Eunus; Azam, Sami (2024). "A Review on Large Language Models: Architectures, Applications, Taxonomies, Open Issues and Challenges". IEEE Access. 12: 26839–26874. Bibcode:2024IEEEA..1226839R. doi:10.1109/access.2024.3365742. ISSN 2169-3536.
- ^ Cite error: The named reference
:13was invoked but never defined (see the help page). - ^ Zhou, Zhihan; Ji, Yanrong; Li, Weijian; Dutta, Pratik; Davuluri, Ramana; Liu, Han (2023), DNABERT-2: Efficient Foundation Model and Benchmark For Multi-Species Genome, arXiv:2306.15006, retrieved 2025-02-21
- ^ a b Mihindukulasuriya, Kathie A.; Mars, Ruben A.T.; Johnson, Abigail J.; Ward, Tonya; Priya, Sambhawa; Lekatz, Heather R.; Kalari, Krishna R.; Droit, Lindsay; Zheng, Tenghao; Blekhman, Ran; D'Amato, Mauro; Farrugia, Gianrico; Knights, Dan; Handley, Scott A.; Kashyap, Purna C. (October 2021). "Multi-Omics Analyses Show Disease, Diet, and Transcriptome Interactions With the Virome". Gastroenterology. 161 (4): 1194–1207.e8. doi:10.1053/j.gastro.2021.06.077. ISSN 0016-5085. PMC 8463486. PMID 34245762.
- ^ Stupak, Aleksandra; Kwiatek, Maciej; Gęca, Tomasz; Kwaśniewska, Anna; Mlak, Radosław; Nawrot, Robert; Goździcka-Józefiak, Anna; Kwaśniewski, Wojciech (2024-10-23). "A Virome and Proteomic Analysis of Placental Microbiota in Pregnancies with and without Fetal Growth Restriction". Cells. 13 (21): 1753. doi:10.3390/cells13211753. ISSN 2073-4409. PMID 39513860.
- ^ a b c Xie, Peiwei; Luo, Mei; Fan, Jiahui; Xiong, Lishou (2024-06-29). "Multiomics Analysis Reveals Gut Virome–Bacteria–Metabolite Interactions and Their Associations with Symptoms in Patients with IBS-D". Viruses. 16 (7): 1054. doi:10.3390/v16071054. ISSN 1999-4915. PMC 11281411. PMID 39066219.
- ^ McCall, Camille; Wu, Huiyun; Miyani, Brijen; Xagoraraki, Irene (October 2020). "Identification of multiple potential viral diseases in a large urban center using wastewater surveillance". Water Research. 184: 116160. Bibcode:2020WatRe.18416160M. doi:10.1016/j.watres.2020.116160. ISSN 0043-1354. PMID 32738707.
- ^ Leifels, Mats; Khalilur Rahman, Omar; Sam, I-Ching; Cheng, Dan; Chua, Feng Jun Desmond; Nainani, Dhiraj; Kim, Se Yeon; Ng, Wei Jie; Kwok, Wee Chiew; Sirikanchana, Kwanrawee; Wuertz, Stefan; Thompson, Janelle; Chan, Yoke Fun (2022-10-30). "The one health perspective to improve environmental surveillance of zoonotic viruses: lessons from COVID-19 and outlook beyond". ISME Communications. 2 (1): 107. Bibcode:2022ISMEC...2..107L. doi:10.1038/s43705-022-00191-8. ISSN 2730-6151. PMC 9618154. PMID 36338866.
