AP-1 transcription factor

| AP-1 Proteins (Fos, ATF, JDP) | |
|---|---|
| Identifiers | |
| Symbol | AP-1 |
| InterPro | IPR000837 |
| Transcription factor Jun | |
|---|---|
| Identifiers | |
| Symbol | Leuzip_Jun |
| InterPro | IPR002112 |
Activator protein 1 (AP-1) is a transcription factor that regulates gene expression in response to a variety of stimuli, including cytokines, growth factors, stress, and bacterial and viral infections.[1] AP-1 controls a number of cellular processes including differentiation, proliferation, and apoptosis.[2] The structure of AP-1 is a heterodimer composed of proteins belonging to the c-Fos, c-Jun, ATF and JDP families.
History
[edit]AP-1 was first discovered as a TPA-activated transcription factor that bound to a cis-regulatory element of the human metallothionein IIa (hMTIIa) promoter and SV40.[3] The AP-1 binding site was identified as the 12-O-Tetradecanoylphorbol-13-acetate (TPA) response element (TRE) with the consensus sequence 5’-TGA G/C TCA-3’.[4] The AP-1 subunit Jun was identified as a novel oncoprotein of avian sarcoma virus, and Fos-associated p39 protein was identified as the transcript of the cellular Jun gene. Fos was first isolated as the cellular homologue of two viral v-fos oncogenes, both of which induce osteosarcoma in mice and rats.[5] Since its discovery, AP-1 has been found to be associated with numerous regulatory and physiological processes, and new relationships are still being investigated.
Structure
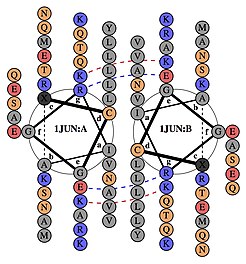
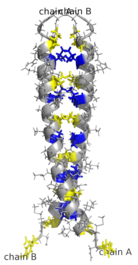
[edit]AP-1 transcription factor is assembled through the dimerization of a characteristic bZIP domain (basic region leucine zipper) in the Fos and Jun subunits. A typical bZIP domain consists of a “leucine zipper” region, and a “basic region”. The leucine zipper is responsible for dimerization of the Jun and Fos protein subunits. This structural motif twists two alpha helical protein domains into a “coiled coil,” characterized by a periodicity of 3.5 residues per turn and repetitive leucines appearing at every seventh position of the polypeptide chain. Due to the amino acid sequence and the periodicity of the helices, the leucine side chains are arranged along one face of the α helix and form a hydrophobic surface that modulates dimerization.[6] Hydrophobic residues additional to leucine also form the characteristic 3-4 repeat of α helices involved in “coiled-coil” interactions, and help contribute to the hydrophobic packing that drives dimerization. Together, this hydrophobic surface holds the two subunits together.[7][8]
The basic region of the bZIP domain is just upstream to the leucine zipper, and contains positively charged residues. This region interacts with DNA target sites.[9] Apart from the “leucine zipper” and the “basic region” which are important for dimerization and DNA-binding, the c-jun protein contains three short regions, which consist of clusters of negatively charged amino acids in its N-terminal half that are important for transcriptional activation in vivo.[10]
Dimerization happens between the products of the c-jun and c-fos protooncogenes, and is required for DNA-binding. Jun proteins can form both homo and heterodimers and therefore are capable of binding to DNA by themselves. However, Fos proteins do not dimerize with each other and therefore can only bind to DNA when bound with Jun.[11][12] The Jun-Fos heterodimer is more stable and has higher DNA-binding activity than Jun homodimers.
Function
[edit]AP-1 transcription factor has been shown to have a hand in a wide range of cellular processes, including cell growth, differentiation, and apoptosis. AP-1 activity is often regulated via post-translational modifications, DNA binding dimer composition, and interaction with various binding partners. AP-1 transcription factors are also associated with numerous physiological functions especially in determination of organisms’ life span and tissue regeneration. Below are some of the other important functions and biological roles AP-1 transcription factors have been shown to be involved in.
Cell growth, proliferation and senescence
[edit]The AP-1 transcription factor has been shown to play numerous roles in cell growth and proliferation. In particular, c-Fos and c-Jun seem to be major players in these processes. C-jun has been shown to be essential for fibroblast proliferation,[13] and levels of both AP-1 subunits have been shown to be expressed above basal levels during cell division.[14] C-fos has also been shown to increase in expression in response to the introduction of growth factors in the cell, further supporting its suggested involvement in the cell cycle. The growth factors TGF alpha, TGF beta, and IL2 have all been shown to stimulate c-Fos, and thereby stimulate cellular proliferation via AP-1 activation.[10]
Cellular senescence has been identified as "a dynamic and reversible process regulated by (in)activation of a predetermined enhancer landscape controlled by the pioneer transcription factor AP-1", which "defines the organizational principles of the transcription factor network that drives the transcriptional programme of senescent cells".[15][16]
Cellular differentiation
[edit]AP-1 transcription is deeply involved in the modulation of gene expression. Changes in cellular gene expression in the initiation of DNA synthesis and the formation of differentiated derivatives can lead to cellular differentiation.[10] AP-1 has been shown to be involved in cell differentiation in several systems. For example, by forming stable heterodimers with c-Jun, the bZIP region of c-Fos increases the binding of c-Jun to target genes whose activation is involved in the differentiation of chicken embryo fibroblasts (CEF).[17] It has also been shown to participate in endoderm specification.[18]
Apoptosis
[edit]AP-1 transcription factor is associated with a broad range of apoptosis related interactions. AP-1 activity is induced by numerous extracellular matrix and genotoxic agents, suggesting involvement in programmed cell death.[2] Many of these stimuli activate the c-Jun N-terminal kinases (JNKs) leading to the phosphorylation of Jun proteins and enhanced transcriptional activity of AP-1 dependent genes.[2] Increases in the levels of Jun and Fos proteins and JNK activity have been reported in scenarios in which cells undergo apoptosis. For example, inactivated c-Jun-ER cells show a normal morphology, while c-Jun-ER activated cells have been shown to be apoptotic.[19]
Regulation of AP-1
[edit]Increased AP-1 levels lead to increased transactivation of target gene expression. Regulation of AP-1 activity is therefore critical for cell function and occurs through specific interactions controlled by dimer-composition, transcriptional and post-translational events, and interaction with accessory proteins.[20]
AP-1 functions are heavily dependent on the specific Fos and Jun subunits contributing to AP-1 dimers.[10] The outcome of AP-1 activation is dependent on the complex combinatorial patterns of AP-1 component dimers.[2] The AP-1 complex binds to a palindromic DNA motif (5’-TGA G/C TCA-3’) to regulate gene expression, but specificity is dependent on the dimer composition of the bZIP subunit.[2]
Physiological relevance
[edit]AP-1 transcription factor has been shown to be involved in skin physiology, specifically in tissue regeneration. The process of skin metabolism is initiated by signals that trigger undifferentiated proliferative cells to undergo cell differentiation. Therefore, activity of AP-1 subunits in response to extracellular signals may be modified under conditions where the balance of keratinocyte proliferation and differentiation has to be rapidly and temporally altered.[21] The AP-1 transcription factor also has been shown to be involved in breast cancer cell growth through multiple mechanisms, including regulation of cyclin D1, E2F factors and their target genes. c-Jun, which is one of the AP-1 subunits, regulates the growth of breast cancer cells. Activated c-Jun is predominantly expressed at the invasive front in breast cancer and is associated with proliferation of breast cells.[22] Due to the AP-1 regulatory functions in cancer cells, AP-1 modulation is studied as a potential strategy for cancer prevention and therapy.[23][24][25]
Regulome
[edit]See also
[edit]- Activator protein
- Immediate early genes – Genes that are rapidly expressed in response to varied stimuli, without needing new proteins to be synthesized, including c-fos and c-jun[76]
- Transcription factor
References
[edit]- ^ Hess J, Angel P, Schorpp-Kistner M (December 2004). "AP-1 subunits: quarrel and harmony among siblings". Journal of Cell Science. 117 (Pt 25): 5965–73. doi:10.1242/jcs.01589. PMID 15564374.
- ^ a b c d e Ameyar M, Wisniewska M, Weitzman JB (August 2003). "A role for AP-1 in apoptosis: the case for and against". Biochimie. 85 (8): 747–52. doi:10.1016/j.biochi.2003.09.006. PMID 14585541.
- ^ Lee W, Haslinger A, Karin M, Tjian R (January 1987). "Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40". Nature. 325 (6102): 368–72. Bibcode:1987Natur.325..368L. doi:10.1038/325368a0. PMID 3027570. S2CID 4314423.
- ^ Angel P, Imagawa M, Chiu R, Stein B, Imbra RJ, Rahmsdorf HJ, Jonat C, Herrlich P, Karin M (June 1987). "Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor". Cell. 49 (6): 729–39. doi:10.1016/0092-8674(87)90611-8. PMID 3034432. S2CID 23154076.
- ^ Wagner EF (April 2001). "AP-1--Introductory remarks". Oncogene. 20 (19): 2334–5. doi:10.1038/sj.onc.1204416. PMID 11402330.
- ^ Landschulz WH, Johnson PF, McKnight SL (June 1988). "The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins". Science. 240 (4860): 1759–64. Bibcode:1988Sci...240.1759L. doi:10.1126/science.3289117. PMID 3289117.
- ^ O'Shea EK, Rutkowski R, Kim PS (January 1989). "Evidence that the leucine zipper is a coiled coil". Science. 243 (4890): 538–42. Bibcode:1989Sci...243..538O. doi:10.1126/science.2911757. PMID 2911757.
- ^ O'Shea EK, Rutkowski R, Stafford WF, Kim PS (August 1989). "Preferential heterodimer formation by isolated leucine zippers from fos and jun". Science. 245 (4918): 646–8. Bibcode:1989Sci...245..646O. doi:10.1126/science.2503872. PMID 2503872.
- ^ Vogt PK, Bos TJ (1990). "jun: oncogene and transcription factor". Advances in Cancer Research. 55: 1–35. doi:10.1016/s0065-230x(08)60466-2. ISBN 9780120066551. PMID 2166997.
- ^ a b c d Angel P, Karin M (December 1991). "The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation". Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1072 (2–3): 129–57. doi:10.1016/0304-419X(91)90011-9. PMID 1751545.
- ^ Kouzarides T, Ziff E (December 1988). "The role of the leucine zipper in the fos-jun interaction". Nature. 336 (6200): 646–51. Bibcode:1988Natur.336..646K. doi:10.1038/336646a0. PMID 2974122. S2CID 4355663.
- ^ Nakabeppu Y, Ryder K, Nathans D (December 1988). "DNA binding activities of three murine Jun proteins: stimulation by Fos". Cell. 55 (5): 907–15. doi:10.1016/0092-8674(88)90146-8. PMID 3142691. S2CID 11057487.
- ^ Karin M, Liu Z, Zandi E (April 1997). "AP-1 function and regulation". Current Opinion in Cell Biology. 9 (2): 240–6. doi:10.1016/S0955-0674(97)80068-3. PMID 9069263.
- ^ Yamashita J, McCauley LK (2006). "The Activating Protein-1 Transcriptional Complex: Essential and Multifaceted Roles in Bone". Clinical Reviews in Bone and Mineral Metabolism. 4 (2): 107–122. doi:10.1385/BMM:4:2:107. S2CID 90318354.
- ^ Zumerle S, Alimonti A (2020). "In and out from senescence". Nat Cell Biol. 22 (7): 753–754. doi:10.1038/s41556-020-0540-x. PMID 32591745. S2CID 220071911.
- ^ Martínez-Zamudio R, Roux P, de Freitas J, et al. (2020). "AP-1 imprints a reversible transcriptional programme of senescent cells". Nat Cell Biol. 22 (7): 842–855. doi:10.1038/s41556-020-0529-5. PMC 7899185. PMID 32514071. S2CID 219543898.
- ^ Shaulian E, Karin M (May 2002). "AP-1 as a regulator of cell life and death". Nature Cell Biology. 4 (5): E131–6. doi:10.1038/ncb0502-e131. PMID 11988758. S2CID 34337538.
- ^ Madrigal P, Deng S, Feng Y, Militi S, Goh KJ, Nibhani R, Grandy R, Osnato A, Ortmann D, Brown S, Pauklin S (January 25, 2023). "Epigenetic and transcriptional regulations prime cell fate before division during human pluripotent stem cell differentiation" (PDF). Nature Communications. 14 (405): 405. Bibcode:2023NatCo..14..405M. doi:10.1038/s41467-023-36116-9. PMC 9876972. PMID 36697417.
- ^ Bossy-Wetzel E, Bakiri L, Yaniv M (April 1997). "Induction of apoptosis by the transcription factor c-Jun". The EMBO Journal. 16 (7): 1695–709. doi:10.1093/emboj/16.7.1695. PMC 1169773. PMID 9130714.
- ^ Vesely PW, Staber PB, Hoefler G, Kenner L (July 2009). "Translational regulation mechanisms of AP-1 proteins". Mutation Research. 682 (1): 7–12. Bibcode:2009MRRMR.682....7V. doi:10.1016/j.mrrev.2009.01.001. PMID 19167516.
- ^ Angel P, Szabowski A, Schorpp-Kistner M (April 2001). "Function and regulation of AP-1 subunits in skin physiology and pathology". Oncogene. 20 (19): 2413–23. doi:10.1038/sj.onc.1204380. PMID 11402337.
- ^ Shen Q, Uray IP, Li Y, Krisko TI, Strecker TE, Kim HT, Brown PH (January 2008). "The AP-1 transcription factor regulates breast cancer cell growth via cyclins and E2F factors". Oncogene. 27 (3): 366–77. doi:10.1038/sj.onc.1210643. PMID 17637753.
- ^ Eferl R, Wagner EF (November 2003). "AP-1: a double-edged sword in tumorigenesis". Nature Reviews. Cancer. 3 (11): 859–68. doi:10.1038/nrc1209. PMID 14668816. S2CID 35328722.
- ^ Tewari D, Nabavi SF, Nabavi SM, Sureda A, Farooqi AA, Atanasov AG, Vacca RA, Sethi G, Bishayee A (February 2018). "Targeting activator protein 1 signaling pathway by bioactive natural agents: Possible therapeutic strategy for cancer prevention and intervention". Pharmacological Research. 128: 366–375. doi:10.1016/j.phrs.2017.09.014. PMID 28951297. S2CID 20160666.
- ^ Kamide D, Yamashita T, Araki K, Tomifuji M, Tanaka Y, Tanaka S, Shiozawa S, Shiotani A (May 2016). "Selective activator protein-1 inhibitor T-5224 prevents lymph node metastasis in an oral cancer model". Cancer Science. 107 (5): 666–73. doi:10.1111/cas.12914. PMC 4970834. PMID 26918517.
- ^ Proffitt J, Crabtree G, Grove M, Daubersies P, Bailleul B, Wright E, Plumb M (January 1995). "An ATF/CREB-binding site is essential for cell-specific and inducible transcription of the murine MIP-1 beta cytokine gene". Gene. 152 (2): 173–9. doi:10.1016/0378-1119(94)00701-S. PMID 7835696.
- ^ Rainio EM, Sandholm J, Koskinen PJ (February 2002). "Cutting edge: Transcriptional activity of NFATc1 is enhanced by the Pim-1 kinase". Journal of Immunology. 168 (4): 1524–7. doi:10.4049/jimmunol.168.4.1524. PMID 11823475.
- ^ Sanyal S, Sandstrom DJ, Hoeffer CA, Ramaswami M (April 2002). "AP-1 functions upstream of CREB to control synaptic plasticity in Drosophila". Nature. 416 (6883): 870–4. Bibcode:2002Natur.416..870S. doi:10.1038/416870a. PMID 11976688. S2CID 4329320.
- ^ Hirayama J, Cardone L, Doi M, Sassone-Corsi P (July 2005). "Common pathways in circadian and cell cycle clocks: light-dependent activation of Fos/AP-1 in zebrafish controls CRY-1a and WEE-1". Proceedings of the National Academy of Sciences of the United States of America. 102 (29): 10194–9. Bibcode:2005PNAS..10210194H. doi:10.1073/pnas.0502610102. PMC 1177375. PMID 16000406.
- ^ Wai PY, Mi Z, Gao C, Guo H, Marroquin C, Kuo PC (July 2006). "Ets-1 and runx2 regulate transcription of a metastatic gene, osteopontin, in murine colorectal cancer cells". The Journal of Biological Chemistry. 281 (28): 18973–82. doi:10.1074/jbc.M511962200. PMID 16670084.
- ^ Collins-Hicok J, Lin L, Spiro C, Laybourn PJ, Tschumper R, Rapacz B, McMurray CT (May 1994). "Induction of the rat prodynorphin gene through Gs-coupled receptors may involve phosphorylation-dependent derepression and activation". Molecular and Cellular Biology. 14 (5): 2837–48. doi:10.1128/MCB.14.5.2837. PMC 358652. PMID 8164647.
- ^ a b Behren A, Simon C, Schwab RM, Loetzsch E, Brodbeck S, Huber E, Stubenrauch F, Zenner HP, Iftner T (December 2005). "Papillomavirus E2 protein induces expression of the matrix metalloproteinase-9 via the extracellular signal-regulated kinase/activator protein-1 signaling pathway". Cancer Research. 65 (24): 11613–21. doi:10.1158/0008-5472.CAN-05-2672. PMID 16357172.
- ^ Hennigan RF, Stambrook PJ (August 2001). "Dominant negative c-jun inhibits activation of the cyclin D1 and cyclin E kinase complexes". Molecular Biology of the Cell. 12 (8): 2352–63. doi:10.1091/mbc.12.8.2352. PMC 58599. PMID 11514621.
- ^ Knöchel S, Schuler-Metz A, Knöchel W (November 2000). "c-Jun (AP-1) activates BMP-4 transcription in Xenopus embryos". Mechanisms of Development. 98 (1–2): 29–36. doi:10.1016/S0925-4773(00)00448-2. PMID 11044605. S2CID 18150052.
- ^ a b Kidd M, Hinoue T, Eick G, Lye KD, Mane SM, Wen Y, Modlin IM (December 2004). "Global expression analysis of ECL cells in Mastomys natalensis gastric mucosa identifies alterations in the AP-1 pathway induced by gastrin-mediated transformation". Physiological Genomics. 20 (1): 131–42. doi:10.1152/physiolgenomics.00216.2003. PMID 15602048.
- ^ Heim JM, Singh S, Fülle HJ, Gerzer R (January 1992). "Comparison of a cloned ANF-sensitive guanylate cyclase (GC-A) with particulate guanylate cyclase from adrenal cortex". Naunyn-Schmiedeberg's Archives of Pharmacology. 345 (1): 64–70. doi:10.1007/BF00175471. PMID 1347156. S2CID 22605840.
- ^ Kuo YR, Wu WS, Wang FS (April 2007). "Flashlamp pulsed-dye laser suppressed TGF-beta1 expression and proliferation in cultured keloid fibroblasts is mediated by MAPK pathway". Lasers in Surgery and Medicine. 39 (4): 358–64. doi:10.1002/lsm.20489. PMID 17457842. S2CID 25556684.
- ^ Wu J, Bresnick EH (March 2007). "Glucocorticoid and growth factor synergism requirement for Notch4 chromatin domain activation". Molecular and Cellular Biology. 27 (6): 2411–22. doi:10.1128/MCB.02152-06. PMC 1820485. PMID 17220278.
- ^ Martins G, Calame K (2008). "Regulation and functions of Blimp-1 in T and B lymphocytes". Annual Review of Immunology. 26: 133–69. doi:10.1146/annurev.immunol.26.021607.090241. PMID 18370921.
- ^ Lunec J, Holloway K, Cooke M, Evans M (2003). "Redox-regulation of DNA repair". BioFactors. 17 (1–4): 315–24. doi:10.1002/biof.5520170131. PMID 12897453. S2CID 30654477.
- ^ Manicassamy S, Gupta S, Huang Z, Sun Z (June 2006). "Protein kinase C-theta-mediated signals enhance CD4+ T cell survival by up-regulating Bcl-xL". Journal of Immunology. 176 (11): 6709–16. doi:10.4049/jimmunol.176.11.6709. PMID 16709830.
- ^ Wang N, Verna L, Hardy S, Forsayeth J, Zhu Y, Stemerman MB (September 1999). "Adenovirus-mediated overexpression of c-Jun and c-Fos induces intercellular adhesion molecule-1 and monocyte chemoattractant protein-1 in human endothelial cells". Arteriosclerosis, Thrombosis, and Vascular Biology. 19 (9): 2078–84. doi:10.1161/01.ATV.19.9.2078. PMID 10479648.
- ^ Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H (May 2008). "miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism". Journal of Molecular Biology. 378 (3): 492–504. doi:10.1016/j.jmb.2008.03.015. PMID 18384814.
- ^ von Knethen A, Callsen D, Brüne B (February 1999). "NF-kappaB and AP-1 activation by nitric oxide attenuated apoptotic cell death in RAW 264.7 macrophages". Molecular Biology of the Cell. 10 (2): 361–72. doi:10.1091/mbc.10.2.361. PMC 25174. PMID 9950682.
- ^ Phelan JP, Millson SH, Parker PJ, Piper PW, Cooke FT (October 2006). "Fab1p and AP-1 are required for trafficking of endogenously ubiquitylated cargoes to the vacuole lumen in S. cerevisiae". Journal of Cell Science. 119 (Pt 20): 4225–34. doi:10.1242/jcs.03188. PMID 17003107.
- ^ Nolasco LH, Turner NA, Bernardo A, Tao Z, Cleary TG, Dong JF, Moake JL (December 2005). "Hemolytic uremic syndrome-associated Shiga toxins promote endothelial-cell secretion and impair ADAMTS13 cleavage of unusually large von Willebrand factor multimers". Blood. 106 (13): 4199–209. doi:10.1182/blood-2005-05-2111. PMC 1895236. PMID 16131569.
- ^ Hommura F, Katabami M, Leaner VD, Donninger H, Sumter TF, Resar LM, Birrer MJ (May 2004). "HMG-I/Y is a c-Jun/activator protein-1 target gene and is necessary for c-Jun-induced anchorage-independent growth in Rat1a cells". Molecular Cancer Research. 2 (5): 305–14. doi:10.1158/1541-7786.305.2.5. PMID 15192124. S2CID 25127065.
- ^ Chang CJ, Chao JC (April 2002). "Effect of human milk and epidermal growth factor on growth of human intestinal Caco-2 cells". Journal of Pediatric Gastroenterology and Nutrition. 34 (4): 394–401. doi:10.1097/00005176-200204000-00015. PMID 11930096. S2CID 25446228.
- ^ Weber JR, Skene JH (July 1998). "The activity of a highly promiscuous AP-1 element can be confined to neurons by a tissue-selective repressive element". The Journal of Neuroscience. 18 (14): 5264–74. doi:10.1523/jneurosci.18-14-05264.1998. PMC 6793474. PMID 9651209.
- ^ Lee W, Mitchell P, Tjian R (June 1987). "Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements". Cell. 49 (6): 741–52. doi:10.1016/0092-8674(87)90612-X. PMID 3034433. S2CID 37036603.
- ^ Cohen MP, Ziyadeh FN, Lautenslager GT, Cohen JA, Shearman CW (May 1999). "Glycated albumin stimulation of PKC-beta activity is linked to increased collagen IV in mesangial cells". The American Journal of Physiology. 276 (5 Pt 2): F684–90. doi:10.1152/ajprenal.1999.276.5.F684. PMID 10330050.
- ^ Stark CJ, Atreya CD (April 2005). "Molecular advances in the cell biology of SARS-CoV and current disease prevention strategies". Virology Journal. 2: 35. doi:10.1186/1743-422X-2-35. PMC 1087510. PMID 15833113.
- ^ a b c Lane SJ, Adcock IM, Richards D, Hawrylowicz C, Barnes PJ, Lee TH (December 1998). "Corticosteroid-resistant bronchial asthma is associated with increased c-fos expression in monocytes and T lymphocytes". The Journal of Clinical Investigation. 102 (12): 2156–64. doi:10.1172/JCI2680. PMC 509170. PMID 9854051.
- ^ Steiner C, Peters WH, Gallagher EP, Magee P, Rowland I, Pool-Zobel BL (March 2007). "Genistein protects human mammary epithelial cells from benzo(a)pyrene-7,8-dihydrodiol-9,10-epoxide and 4-hydroxy-2-nonenal genotoxicity by modulating the glutathione/glutathione S-transferase system". Carcinogenesis. 28 (3): 738–48. doi:10.1093/carcin/bgl180. PMID 17065199.
- ^ Ahn JD, Morishita R, Kaneda Y, Lee KU, Park JY, Jeon YJ, Song HS, Lee IK (June 2001). "Transcription factor decoy for activator protein-1 (AP-1) inhibits high glucose- and angiotensin II-induced type 1 plasminogen activator inhibitor (PAI-1) gene expression in cultured human vascular smooth muscle cells". Diabetologia. 44 (6): 713–20. doi:10.1007/s001250051680. PMID 11440364.
- ^ Kang S, Fisher GJ, Voorhees JJ (November 2001). "Photoaging: pathogenesis, prevention, and treatment". Clinics in Geriatric Medicine. 17 (4): 643–59, v–vi. doi:10.1016/S0749-0690(05)70091-4. PMID 11535421.
- ^ Navasa M, Gordon DA, Hariharan N, Jamil H, Shigenaga JK, Moser A, Fiers W, Pollock A, Grunfeld C, Feingold KR (June 1998). "Regulation of microsomal triglyceride transfer protein mRNA expression by endotoxin and cytokines". Journal of Lipid Research. 39 (6): 1220–30. doi:10.1016/S0022-2275(20)32546-3. PMID 9643353.
- ^ Suetsugu M, Takano A, Nagai A, Takeshita A, Hirose K, Matsumoto K, et al. (2007). "Retinoic acid inhibits serum-stimulated activator protein-1 via suppression of c-fos and c-jun gene expressions during the vitamin-induced differentiation of mouse osteoblastic cell line MC3T3-E1 cells" (PDF). J. Meikai Dent. Med. 36 (1): 42–50.
- ^ Inagi R, Miyata T, Nangaku M, Ueyama H, Takeyama K, Kato S, Kurokawa K (November 2002). "Transcriptional regulation of a mesangium-predominant gene, megsin". Journal of the American Society of Nephrology. 13 (11): 2715–22. doi:10.1097/01.ASN.0000033507.32175.FA. PMID 12397041.
- ^ Kim S, Yu SS, Lee IS, Ohno S, Yim J, Kim S, Kang HS (April 1999). "Human cytomegalovirus IE1 protein activates AP-1 through a cellular protein kinase(s)". The Journal of General Virology. 80 ( Pt 4) (4): 961–9. doi:10.1099/0022-1317-80-4-961. PMID 10211966.
- ^ Masuda A, Yoshikai Y, Kume H, Matsuguchi T (November 2004). "The interaction between GATA proteins and activator protein-1 promotes the transcription of IL-13 in mast cells". Journal of Immunology. 173 (9): 5564–73. doi:10.4049/jimmunol.173.9.5564. PMID 15494506.
- ^ Navas TA, Baldwin DT, Stewart TA (November 1999). "RIP2 is a Raf1-activated mitogen-activated protein kinase kinase". The Journal of Biological Chemistry. 274 (47): 33684–90. doi:10.1074/jbc.274.47.33684. PMID 10559258.
- ^ Simantov R (August 1995). "Neurotransporters: regulation, involvement in neurotoxicity, and the usefulness of antisense nucleic acids". Biochemical Pharmacology. 50 (4): 435–42. doi:10.1016/0006-2952(95)00068-B. PMID 7646547.
- ^ Yang HS, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, Colburn NH (February 2001). "A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation". Oncogene. 20 (6): 669–76. doi:10.1038/sj.onc.1204137. PMID 11314000.
- ^ Xie J, Pan H, Yoo S, Gao SJ (December 2005). "Kaposi's sarcoma-associated herpesvirus induction of AP-1 and interleukin 6 during primary infection mediated by multiple mitogen-activated protein kinase pathways". Journal of Virology. 79 (24): 15027–37. doi:10.1128/JVI.79.24.15027-15037.2005. PMC 1316010. PMID 16306573.
- ^ Khan MA, Bouzari S, Ma C, Rosenberger CM, Bergstrom KS, Gibson DL, Steiner TS, Vallance BA (April 2008). "Flagellin-dependent and -independent inflammatory responses following infection by enteropathogenic Escherichia coli and Citrobacter rodentium". Infection and Immunity. 76 (4): 1410–22. doi:10.1128/IAI.01141-07. PMC 2292885. PMID 18227166.
- ^ Kida Y, Inoue H, Shimizu T, Kuwano K (January 2007). "Serratia marcescens serralysin induces inflammatory responses through protease-activated receptor 2". Infection and Immunity. 75 (1): 164–74. doi:10.1128/IAI.01239-06. PMC 1828393. PMID 17043106.
- ^ Gutzman JH, Rugowski DE, Schroeder MD, Watters JJ, Schuler LA (December 2004). "Multiple kinase cascades mediate prolactin signals to activating protein-1 in breast cancer cells". Molecular Endocrinology. 18 (12): 3064–75. doi:10.1210/me.2004-0187. PMC 1634796. PMID 15319452.
- ^ Brinkmann MM, Glenn M, Rainbow L, Kieser A, Henke-Gendo C, Schulz TF (September 2003). "Activation of mitogen-activated protein kinase and NF-kappaB pathways by a Kaposi's sarcoma-associated herpesvirus K15 membrane protein". Journal of Virology. 77 (17): 9346–58. doi:10.1128/JVI.77.17.9346-9358.2003. PMC 187392. PMID 12915550.
- ^ Greenstein S, Ghias K, Krett NL, Rosen ST (June 2002). "Mechanisms of glucocorticoid-mediated apoptosis in hematological malignancies". Clinical Cancer Research. 8 (6): 1681–94. PMID 12060604.
- ^ Yokoo T, Kitamura M (May 1996). "Antioxidant PDTC induces stromelysin expression in mesangial cells via a tyrosine kinase-AP-1 pathway". The American Journal of Physiology. 270 (5 Pt 2): F806–11. doi:10.1152/ajprenal.1996.270.5.F806. PMID 8928842.
- ^ Chang CF, Cho S, Wang J (April 2014). "(-)-Epicatechin protects hemorrhagic brain via synergistic Nrf2 pathways". Annals of Clinical and Translational Neurology. 1 (4): 258–271. doi:10.1002/acn3.54. PMC 3984761. PMID 24741667.
- ^ Gibbings DJ, Ghetu AF, Dery R, Befus AD (February 2008). "Macrophage migration inhibitory factor has a MHC class I-like motif and function". Scandinavian Journal of Immunology. 67 (2): 121–32. doi:10.1111/j.1365-3083.2007.02046.x. PMID 18201367.
- ^ Uniprot Database
- ^ Hseu YC, Vudhya Gowrisankar Y, Chen XZ, Yang YC, Yang HL (Feb 2020). "The antiaging activity of ergothioneine in UVA-irradiated human dermal fibroblasts via the inhibition of the AP-1 pathway and the activation of Nrf2-mediated antioxidant genes". Oxid Med Cell Longev. 2020 (2576823): 1–13. doi:10.1155/2020/2576823. PMC 7038158. PMID 32104530.
- ^ Bahrami S, Drabløs F (2016). "Gene regulation in the immediate-early response process". Advances in Biological Regulation. 62: 37–49. doi:10.1016/j.jbior.2016.05.001. PMID 27220739.