User:Emsci1013/sandbox
The homeobox was discovered independently in 1984 by Ernst Hafen, Michael Levine, and William McGinnis working in the lab of Walter Jakob Gehring at the University of Basel, Switzerland; and by Matthew P. Scott and Amy Weiner, who were then working with Thomas Kaufman at Indiana University in Bloomington. [1][2]
The existence of genes sharing the homeobox sequence was independently reported by Ernst Hafen, Michael Levine, William McGinnis, and Walter Jakob Gehring of the University of Basel in Switzerland and Matthew P. Scott and Amy Weiner of Indiana University in Bloomington in 1984.
The existence of homeobox genes was first discovered in Drosophila by isolating the gene responsible for a homeotic transformation where legs grow from the head instead of the expected antennae. Walter Gehring identified a gene called antennapedia that caused this homeotic phenotype.[3] Analysis of antennapedia revealed that this gene contained a 180 base pair sequence that encoded a DNA binding domain, which Gehring termed the "homeobox".[4] Isolation of homologous genes by Edward de Robertis and Willian McGinnis revealed that numerous genes from a variety of species contained the homeobox.[5][6] Subsequent phylogenetic studies detailing the evolutionary relationship between homeobox-containing genes showed that these genes are present in all bilaterian animals.
Homeobox genes
[edit]Homeobox genes are a super-family of genes that contain the homeobox DNA sequence. Homeobox genes encode homeodomain proteins that act as transcription factors. There are
Regulation
[edit]Homeobox genes are regulated
Colinearity
[edit]
Biological function
[edit]Homeodomain proteins function as transcription factors due to the DNA binding properties of the conserved HTH motif. Homeodomain proteins are considered to be master control genes, meaning that a single protein can regulate expression of many target genes. Homeodomain proteins direct the formation of the body axes and body structures during early embryonic development.[7] Many homeodomain proteins induce cellular differentiation by initiating the cascades of coregulated genes required to produce individual tissues and organs. Other proteins in the family, such as NANOG are involved in maintaining pluripotency and preventing cell differentiation.
Regulation
[edit]Hox genes and their associated microRNAs are highly conserved developmental master regulators with tight tissue-specific, spatiotemporal control. These genes are known to be dysregulated in several cancers and are often controlled by DNA methylation.[8][9] The regulation of Hox genes is highly complex and involves reciprocal interactions, mostly inhibitory. Drosophila is known to use the Polycomb and Trithorax Complexes to maintain the expression of Hox genes after the down-regulation of the pair-rule and gap genes that occurs during larval development. Polycomb-group proteins can silence the Hox genes by modulation of chromatin structure.[10]
Mutations
[edit]Mutations to homeobox genes can produce easily visible phenotypic changes in body segment identity, such as the Antennapedia and Bithorax mutant phenotypes in Drosophila. Duplication of homeobox genes can produce new body segments, and such duplications are likely to have been important in the evolution of segmented animals.
Types of homeobox genes
[edit]Hox genes
[edit]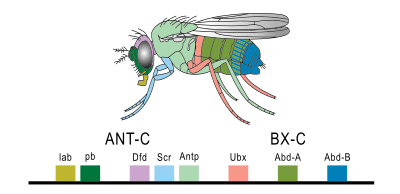
Hox genes are the most commonly known subset of homeobox genes. They are essential metazoan genes that determine the identity of embryonic regions along the anterior-posterior axis.[11] The first vertebrate Hox gene was isolated in Xenopus by Edward De Robertis and colleagues in 1984.[12] The main interest in this set of genes stems from their unique behavior and arrangement in the genome. Hox genes are typically found in an organized cluster. The linear order of Hox genes within a cluster is directly correlated to the order they are expressed in both time and space during development. This phenomenon is called colinearity.
Mutations in these homeotic genes cause displacement of body segments during embryonic development. This is called ectopia. For example, when one gene is lost the segment develops into a more anterior one, while a mutation that leads to a gain of function causes a segment to develop into a more posterior one. Famous examples are Antennapedia and bithorax in Drosophila, which can cause the development of legs instead of antennae and the development of a duplicated thorax, respectively.[13]
In vertebrates, the four paralog clusters are partially redundant in function, but have also acquired several derived functions. For example, HoxA and HoxD specify segment identity along the limb axis.[14][15] Specific members of the Hox family have been implicated in vascular remodeling, angiogenesis, and disease by orchestrating changes in matrix degradation, integrins, and components of the ECM.[16] HoxA5 is implicated in atherosclerosis.[8][17] HoxD3 and HoxB3 are proinvasive, angiogenic genes that upregulate b3 and a5 integrins and Efna1 in ECs, respectively.[18][19][20][21] HoxA3 induces endothelial cell (EC) migration by upregulating MMP14 and uPAR. Conversely, HoxD10 and HoxA5 have the opposite effect of suppressing EC migration and angiogenesis, and stabilizing adherens junctions by upregulating TIMP1/downregulating uPAR and MMP14, and by upregulating Tsp2/downregulating VEGFR2, Efna1, Hif1alpha and COX-2, respectively.[22][23] HoxA5 also upregulates the tumor suppressor p53 and Akt1 by downregulation of PTEN.[24] Suppression of HoxA5 has been shown to attenuate hemangioma growth.[25] HoxA5 has far-reaching effects on gene expression, causing ~300 genes to become upregulated upon its induction in breast cancer cell lines.[25] HoxA5 protein transduction domain overexpression prevents inflammation shown by inhibition of TNFalpha-inducible monocyte binding to HUVECs.[26][27][28]
NKL genes
[edit]LIM genes
[edit]LIM genes encode two 60 amino acid cysteine and histidine-rich LIM domains and a homeodomain. The LIM domains function in protein-protein interactions and can bind zinc molecules. LIM domain proteins are found in both the cytosol and the nucleus, and function in cytoskeletal remodeling, at focal adhesion sites, as scaffolds for protein complexes, and as transcription factors.
Pax genes
[edit]Most Pax genes contain a homeobox and a paired domain that also binds DNA to increase binding specificity, though some Pax genes have lost all or part of the homeobox sequence.[29] Pax genes function in embryo segmentation, nervous system development, generation of the eye field, skeletal development, and formation of face structures. Pax 6 is a master regulator of eye development, such that the gene is necessary for development of the optic vesicle and subsequent eye structures.[30]
POU genes
[edit]Proteins containing a POU region consist of a homeodomain and a separate, structurally homologous POU domain that contains two helix-turn-helix motifs and also binds DNA. The two domains are linked by a flexible loop that is long enough to stretch around the DNA helix, allowing the two domains to bind on opposite sides of the target DNA, collectively covering an eight-base segment with consensus sequence 5'-ATGCAAAT-3'. The individual domains of POU proteins bind DNA only weakly, but have strong sequence-specific affinity when linked. The POU domain itself has significant structural similarity with repressors expressed in bacteriophages, particularly lambda phage.
POU genes function in
PRD genes
[edit]
SINE genes
[edit]
 | This is a user sandbox of Emsci1013. You can use it for testing or practicing edits. This is not the sandbox where you should draft your assigned article for a dashboard.wikiedu.org course. To find the right sandbox for your assignment, visit your Dashboard course page and follow the Sandbox Draft link for your assigned article in the My Articles section. |
- ^ McGinnis, W.; Levine, M. S.; Hafen, E.; Kuroiwa, A.; Gehring, W. J. (1984). "A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes". Nature. 308 (5958): 428–433. doi:10.1038/308428a0. ISSN 1476-4687. PMID 6323992. S2CID 4235713.
- ^ Scott, M. P.; Weiner, A. J. (1984-07-01). "Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila". Proceedings of the National Academy of Sciences. 81 (13): 4115–4119. doi:10.1073/pnas.81.13.4115. ISSN 0027-8424. PMC 345379. PMID 6330741.
- ^ Garber, R L; Kuroiwa, A; Gehring, W J (1983). "Genomic and cDNA clones of the homeotic locus Antennapedia in Drosophila". The EMBO Journal. 2 (11): 2027–2036. doi:10.1002/j.1460-2075.1983.tb01696.x. ISSN 0261-4189. PMC 555405. PMID 6416827.
- ^ "Walter Jakob Gehring (1939-2014) | The Embryo Project Encyclopedia". embryo.asu.edu. Retrieved 2019-12-09.
- ^ Carrasco, Andrés E.; McGinnis, William; Gehring, Walter J.; De Robertis, Eddy M. (1984). "Cloning of an X. laevis gene expressed during early embryogenesis coding for a peptide region homologous to Drosophila homeotic genes". Cell. 37 (2): 409–414. doi:10.1016/0092-8674(84)90371-4. PMID 6327066. S2CID 30114443.
- ^ McGinnis, William; Garber, Richard L.; Wirz, Johannes; Kuroiwa, Atsushi; Gehring, Walter J. (1984-06-01). "A homologous protein-coding sequence in drosophila homeotic genes and its conservation in other metazoans". Cell. 37 (2): 403–408. doi:10.1016/0092-8674(84)90370-2. ISSN 0092-8674. PMID 6327065. S2CID 40456645.
- ^ Corsetti MT, Briata P, Sanseverino L, Daga A, Airoldi I, Simeone A, Palmisano G, Angelini C, Boncinelli E, Corte G (September 1992). "Differential DNA binding properties of three human homeodomain proteins". Nucleic Acids Research. 20 (17): 4465–72. doi:10.1093/nar/20.17.4465. PMC 334173. PMID 1357628.
- ^ a b Dunn J, Thabet S, Jo H (July 2015). "Flow-Dependent Epigenetic DNA Methylation in Endothelial Gene Expression and Atherosclerosis". Arteriosclerosis, Thrombosis, and Vascular Biology. 35 (7): 1562–9. doi:10.1161/ATVBAHA.115.305042. PMC 4754957. PMID 25953647.
- ^ Bhatlekar S, Fields JZ, Boman BM (August 2014). "HOX genes and their role in the development of human cancers". Journal of Molecular Medicine. 92 (8): 811–23. doi:10.1007/s00109-014-1181-y. PMID 24996520. S2CID 17159381.
- ^ Portoso M, Cavalli G (2008). "The Role of RNAi and Noncoding RNAs in Polycomb Mediated Control of Gene Expression and Genomic Programming". RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity. Caister Academic Press. ISBN 978-1-904455-25-7.
- ^ Alonso CR (November 2002). "Hox proteins: sculpting body parts by activating localized cell death". Current Biology. 12 (22): R776-8. doi:10.1016/S0960-9822(02)01291-5. PMID 12445403. S2CID 17558233.
- ^ Carrasco AE, McGinnis W, Gehring WJ, De Robertis EM (June 1984). "Cloning of an X. laevis gene expressed during early embryogenesis coding for a peptide region homologous to Drosophila homeotic genes". Cell. 37 (2): 409–14. doi:10.1016/0092-8674(84)90371-4. PMID 6327066. S2CID 30114443.
- ^ Schneuwly, Stephan; Klemenz, Roman; Gehring, Walter J. (1987). "Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia". Nature. 325 (6107): 816–818. doi:10.1038/325816a0. ISSN 1476-4687. PMID 3821869. S2CID 4320668.
- ^ Fromental-Ramain, C.; Warot, X.; Messadecq, N.; LeMeur, M.; Dollé, P.; Chambon, P. (1996). "Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod". Development. 122 (10): 2997–3011. doi:10.1242/dev.122.10.2997. ISSN 0950-1991. PMID 8898214.
- ^ Zákány, József; Duboule, Denis (1999-03-29). "Hox genes in digit development and evolution". Cell and Tissue Research. 296 (1): 19–25. doi:10.1007/s004410051262. ISSN 0302-766X. PMID 10199961. S2CID 3192774.
- ^ Gorski DH, Walsh K (November 2000). "The role of homeobox genes in vascular remodeling and angiogenesis". Circulation Research. 87 (10): 865–72. doi:10.1161/01.res.87.10.865. PMID 11073881. S2CID 11982238.
- ^ Dunn J, Simmons R, Thabet S, Jo H (October 2015). "The role of epigenetics in the endothelial cell shear stress response and atherosclerosis". The International Journal of Biochemistry & Cell Biology. 67: 167–76. doi:10.1016/j.biocel.2015.05.001. PMC 4592147. PMID 25979369.
- ^ Boudreau N, Andrews C, Srebrow A, Ravanpay A, Cheresh DA (October 1997). "Induction of the angiogenic phenotype by Hox D3". The Journal of Cell Biology. 139 (1): 257–64. doi:10.1083/jcb.139.1.257. PMC 2139816. PMID 9314544.
- ^ Boudreau NJ, Varner JA (February 2004). "The homeobox transcription factor Hox D3 promotes integrin alpha5beta1 expression and function during angiogenesis". The Journal of Biological Chemistry. 279 (6): 4862–8. doi:10.1074/jbc.M305190200. PMID 14610084.
- ^ Myers C, Charboneau A, Boudreau N (January 2000). "Homeobox B3 promotes capillary morphogenesis and angiogenesis". The Journal of Cell Biology. 148 (2): 343–51. doi:10.1083/jcb.148.2.343. PMC 2174277. PMID 10648567.
- ^ Chen Y, Xu B, Arderiu G, Hashimoto T, Young WL, Boudreau N, Yang GY (November 2004). "Retroviral delivery of homeobox D3 gene induces cerebral angiogenesis in mice". Journal of Cerebral Blood Flow and Metabolism. 24 (11): 1280–7. doi:10.1097/01.WCB.0000141770.09022.AB. PMID 15545924. S2CID 45880928.
- ^ Myers C, Charboneau A, Cheung I, Hanks D, Boudreau N (December 2002). "Sustained expression of homeobox D10 inhibits angiogenesis". The American Journal of Pathology. 161 (6): 2099–109. doi:10.1016/S0002-9440(10)64488-4. PMC 1850921. PMID 12466126.
- ^ Mace KA, Hansen SL, Myers C, Young DM, Boudreau N (June 2005). "HOXA3 induces cell migration in endothelial and epithelial cells promoting angiogenesis and wound repair". Journal of Cell Science. 118 (Pt 12): 2567–77. doi:10.1242/jcs.02399. PMID 15914537. S2CID 9964731.
- ^ Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N (2005). "A role for Hox A5 in regulating angiogenesis and vascular patterning". Lymphatic Research and Biology. 3 (4): 240–52. doi:10.1089/lrb.2005.3.240. PMID 16379594.
- ^ a b Arderiu G, Cuevas I, Chen A, Carrio M, East L, Boudreau NJ (2007). "HoxA5 stabilizes adherens junctions via increased Akt1". Cell Adhesion & Migration. 1 (4): 185–95. doi:10.4161/cam.1.4.5448. PMC 2634105. PMID 19262140.
- ^ Zhu Y, Cuevas IC, Gabriel RA, Su H, Nishimura S, Gao P, Fields A, Hao Q, Young WL, Yang GY, Boudreau NJ (June 2009). "Restoring transcription factor HoxA5 expression inhibits the growth of experimental hemangiomas in the brain". Journal of Neuropathology and Experimental Neurology. 68 (6): 626–32. doi:10.1097/NEN.0b013e3181a491ce. PMC 2728585. PMID 19458547.
- ^ Chen H, Rubin E, Zhang H, Chung S, Jie CC, Garrett E, Biswal S, Sukumar S (May 2005). "Identification of transcriptional targets of HOXA5". The Journal of Biological Chemistry. 280 (19): 19373–80. doi:10.1074/jbc.M413528200. PMID 15757903.
- ^ Lee JY, Park KS, Cho EJ, Joo HK, Lee SK, Lee SD, Park JB, Chang SJ, Jeon BH (July 2011). "Human HOXA5 homeodomain enhances protein transduction and its application to vascular inflammation". Biochemical and Biophysical Research Communications. 410 (2): 312–6. doi:10.1016/j.bbrc.2011.05.139. PMID 21664342.
- ^ Gruss, Peter; Walther, Claudia (1992-05-29). "Pax in development". Cell. 69 (5): 719–722. doi:10.1016/0092-8674(92)90281-G. ISSN 0092-8674. PMID 1591773. S2CID 44613005.
- ^ Walther, C.; Gruss, P. (1991). "Pax-6, a murine paired box gene, is expressed in the developing CNS". Development (Cambridge, England). 113 (4): 1435–1449. doi:10.1242/dev.113.4.1435. ISSN 0950-1991. PMID 1687460.
