Matching (graph theory)
In the mathematical discipline of graph theory, a matching or independent edge set in a graph is a set of edges without common vertices. It may also be an entire graph consisting of edges without common vertices. Bipartite matching is a special case of a network flow problem.
Definition
Given a graph G = (V,E), a matching M in G is a set of pairwise non-adjacent edges; that is, no two edges share a common vertex.
A vertex is matched (or saturated) if it is an endpoint of one of the edges in the matching. Otherwise the vertex is unmatched.
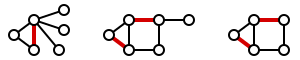
A maximal matching is a matching M of a graph G with the property that if any edge not in M is added to M, it is no longer a matching, that is, M is maximal if it is not a subset of any other matching in graph G. In other words, a matching M of a graph G is maximal if every edge in G has a non-empty intersection with at least one edge in M. The following figure shows examples of maximal matchings (red) in three graphs.
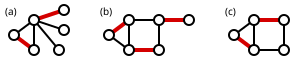
A maximum matching (also known as maximum-cardinality matching[1]) is a matching that contains the largest possible number of edges. There may be many maximum matchings. The matching number of a graph is the size of a maximum matching. Note that every maximum matching is maximal, but not every maximal matching is a maximum matching. The following figure shows examples of maximum matchings in the same three graphs.
A perfect matching (a.k.a. 1-factor) is a matching which matches all vertices of the graph. That is, every vertex of the graph is incident to exactly one edge of the matching. Figure (b) above is an example of a perfect matching. Every perfect matching is maximum and hence maximal. In some literature, the term complete matching is used. In the above figure, only part (b) shows a perfect matching. A perfect matching is also a minimum-size edge cover. Thus, ν(G) ≤ ρ(G) , that is, the size of a maximum matching is no larger than the size of a minimum edge cover.
A near-perfect matching is one in which exactly one vertex is unmatched. This can only occur when the graph has an odd number of vertices, and such a matching must be maximum. In the above figure, part (c) shows a near-perfect matching. If, for every vertex in a graph, there is a near-perfect matching that omits only that vertex, the graph is also called factor-critical.
Given a matching M,
- an alternating path is a path that begins with an unmatched vertex and is a [2] path in which the edges belong alternatively to the matching and not to the matching.
- an augmenting path is an alternating path that starts from and ends on free (unmatched) vertices.
One can prove that a matching is maximum if and only if it does not have any augmenting path. (This result is sometimes called Berge's lemma.)
Properties
In any graph without isolated vertices, the sum of the matching number and the edge covering number equals the number of vertices.[3] If there is a perfect matching, then both the matching number and the edge cover number are |V| / 2.
If A and B are two maximal matchings, then |A| ≤ 2|B| and |B| ≤ 2|A|. To see this, observe that each edge in B \ A can be adjacent to at most two edges in A \ B because A is a matching; moreover each edge in A \ B is adjacent to an edge in B \ A by maximality of B, hence
Further we deduce that
In particular, this shows that any maximal matching is a 2-approximation of a maximum matching and also a 2-approximation of a minimum maximal matching. This inequality is tight: for example, if G is a path with 3 edges and 4 nodes, the size of a minimum maximal matching is 1 and the size of a maximum matching is 2.
Matching polynomials
A generating function of the number of k-edge matchings in a graph is called a matching polynomial. Let G be a graph and mk be the number of k-edge matchings. One matching polynomial of G is
Another definition gives the matching polynomial as
where n is the number of vertices in the graph. Each type has its uses; for more information see the article on matching polynomials.
Algorithms and computational complexity
In unweighted bipartite graphs
Matching problems are often concerned with bipartite graphs. Finding a maximum bipartite matching[4] (often called a maximum cardinality bipartite matching) in a bipartite graph is perhaps the simplest problem.
The Ford–Fulkerson algorithm finds it by repeatedly finding an augmenting path from some x ∈ X to some y ∈ Y and updating the matching M by taking the symmetric difference of that path with M (assuming such a path exists). As each path can be found in time, the running time is . This solution is equivalent to adding a super source with edges to all vertices in , and a super sink with edges from all vertices in , and finding a maximal flow from to . All edges with flow from to then constitute a maximum matching.
An improvement over this is the Hopcroft–Karp algorithm, which runs in time. An alternative randomized approach is based on the fast matrix multiplication algorithm and gives complexity,[5] which is better in theory for sufficiently dense graphs, but in practice the algorithm is slower.[6] Finally, for sparse graphs, is possible with Madry's algorithm based on electric flows.[7]
In addition, the algorithm of Chandran and Hochbaum[6] runs in time that depends on the size of the maximum matching , which for is . Using boolean operations on words of size the complexity is further improved to .
In weighted bipartite graphs
In a weighted bipartite graph, each edge has an associated value. A maximum weighted bipartite matching[4] is defined as a matching where the sum of the values of the edges in the matching have a maximal value. If the graph is not complete bipartite, missing edges are inserted with value zero. Finding such a matching is known as the assignment problem. The Hungarian algorithm solves the assignment problem and it was one of the beginnings of combinatorial optimization algorithms. It uses a modified shortest path search in the augmenting path algorithm. If the Bellman–Ford algorithm is used for this step, the running time of the Hungarian algorithm becomes , or the edge cost can be shifted with a potential to achieve running time with the Dijkstra algorithm and Fibonacci heap.[8]
In general graphs
There is a time algorithm to find a maximum matching or a maximum weight matching in a graph that is not bipartite; it is due to Jack Edmonds, is called the paths, trees, and flowers method or simply Edmonds' algorithm, and uses bidirected edges. A generalization of the same technique can also be used to find maximum independent sets in claw-free graphs. Edmonds' algorithm has subsequently been improved to run in time O(√VE) time, matching the time for bipartite maximum matching.[9]
Another (randomized) algorithm by Mucha and Sankowski,[5] based on the fast matrix multiplication algorithm, gives complexity.
Maximal matchings
A maximal matching can be found with a simple greedy algorithm. A maximum matching is also a maximal matching, and hence it is possible to find a largest maximal matching in polynomial time. However, no polynomial-time algorithm is known for finding a minimum maximal matching, that is, a maximal matching that contains the smallest possible number of edges.
Note that a maximal matching with k edges is an edge dominating set with k edges. Conversely, if we are given a minimum edge dominating set with k edges, we can construct a maximal matching with k edges in polynomial time. Therefore, the problem of finding a minimum maximal matching is essentially equal to the problem of finding a minimum edge dominating set.[10] Both of these two optimisation problems are known to be NP-hard; the decision versions of these problems are classical examples of NP-complete problems.[11] Both problems can be approximated within factor 2 in polynomial time: simply find an arbitrary maximal matching M.[12]
Counting problems
The number of matchings in a graph is known as the Hosoya index of the graph. It is #P-complete to compute this quantity, even for bipartite graphs.[13] It is also #P-complete to count perfect matchings, even in bipartite graphs, because computing the permanent of an arbitrary 0–1 matrix (another #P-complete problem) is the same as computing the number of perfect matchings in the bipartite graph having the given matrix as its biadjacency matrix. However, there exists a fully polynomial time randomized approximation scheme for counting the number of bipartite matchings.[14] A remarkable theorem of Kasteleyn states that the number of perfect matchings in a planar graph can be computed exactly in polynomial time via the FKT algorithm.
The number of perfect matchings in a complete graph Kn (with n even) is given by the double factorial (n − 1)!!.[15] The numbers of matchings in complete graphs, without constraining the matchings to be perfect, are given by the telephone numbers.[16]
Finding all maximally-matchable edges
One of the basic problems in matching theory is to find in a given graph all edges that may be extended to a maximum matching in the graph. (Such edges are called maximally-matchable edges, or allowed edges.) The best deterministic algorithm for solving this problem in general graphs runs in time .[17] There exists a randomized algorithm that solves this problem in time .[18] In the case of bipartite graphs, it is possible to find a single maximum matching and then use it in order to find all maximally-matchable edges in linear time;[19] the resulting overall runtime is for general bipartite graphs and for dense bipartite graphs with . In cases where one of the maximum matchings is known upfront,[20] the overall runtime of the algorithm is .
Characterizations and notes
König's theorem states that, in bipartite graphs, the maximum matching is equal in size to the minimum vertex cover. Via this result, the minimum vertex cover, maximum independent set, and maximum vertex biclique problems may be solved in polynomial time for bipartite graphs.
Hall's marriage theorem provides a characterization of bipartite graphs which have a perfect matching and the Tutte theorem provides a characterization for arbitrary graphs.
A perfect matching is a spanning 1-regular subgraph, a.k.a. a 1-factor. In general, a spanning k-regular subgraph is a k-factor.
Applications
Matching in general graphs
- A Kekulé structure of an aromatic compound consists of a perfect matching of its carbon skeleton, showing the locations of double bonds in the chemical structure. These structures are named after Friedrich August Kekulé von Stradonitz, who showed that benzene (in graph theoretical terms, a 6-vertex cycle) can be given such a structure.[21]
- The Hosoya index is the number of non-empty matchings plus one; it is used in computational chemistry and mathematical chemistry investigations for organic compounds.
Matching in bipartite graphs
- Graduation problem is about choosing minimum set of classes from given requirements for graduation.
- Hitchcock Transport Problem involves bipartite matching as sub-problem.
- Subtree isomorphism problem involves bipartite matching as sub-problem.
See also
- Dulmage–Mendelsohn decomposition, a partition of the vertices of a bipartite graph into subsets such that each edge belongs to a perfect matching if and only if its endpoints belong to the same subset
- Edge coloring, a partition of the edges of a graph into matchings
- Matching preclusion, the minimum number of edges to delete to prevent a perfect matching from existing
- Rainbow matching, a matching in an edge-colored bipartite graph with no repeated colors
- Skew-symmetric graph, a type of graph that can be used to model alternating path searches for matchings
- Stable matching, a matching in which no two elements prefer each other to their matched partners
- Vertex independent set, a set of vertices (rather than edges) no two of which are adjacent to each other
- Stable marriage problem (also known as stable matching problem)
References
- ^ Alan Gibbons, Algorithmic Graph Theory, Cambridge University Press, 1985, Chapter 5.
- ^ http://diestel-graph-theory.com/basic.html
- ^ Gallai, Tibor (1959), "Über extreme Punkt- und Kantenmengen", Ann. Univ. Sci. Budapest. Eötvös Sect. Math., 2: 133–138.
- ^ a b West, Douglas Brent (1999), Introduction to Graph Theory (2nd ed.), Prentice Hall, Chapter 3, ISBN 0-13-014400-2
- ^ a b Mucha, M.; Sankowski, P. (2004), "Maximum Matchings via Gaussian Elimination" (PDF), Proc. 45th IEEE Symp. Foundations of Computer Science, pp. 248–255
- ^ a b Chandran, Bala G.; Hochbaum, Dorit S. (2011), Practical and theoretical improvements for bipartite matching using the pseudoflow algorithm, arXiv:1105.1569,
the theoretically efficient algorithms listed above tend to perform poorly in practice
. - ^ Madry, A (2013), "Navigating Central Path with Electrical Flows: From Flows to Matchings, and Back", Foundations of Computer Science (FOCS), 2013 IEEE 54th Annual Symposium on, pp. 253–262, arXiv:1307.2205
- ^ Fredman, Michael L.; Tarjan, Robert Endre (1987), "Fibonacci heaps and their uses in improved network optimization algorithms", Journal of the ACM, 34 (3): 596–615, doi:10.1145/28869.28874
- ^ Micali, S.; Vazirani, V. V. (1980), "An algorithm for finding maximum matching in general graphs", Proc. 21st IEEE Symp. Foundations of Computer Science, pp. 17–27, doi:10.1109/SFCS.1980.12.
- ^ Yannakakis, Mihalis; Gavril, Fanica (1980), "Edge dominating sets in graphs", SIAM Journal on Applied Mathematics, 38 (3): 364–372, doi:10.1137/0138030.
- ^ Garey, Michael R.; Johnson, David S. (1979), Computers and Intractability: A Guide to the Theory of NP-Completeness, W.H. Freeman, ISBN 0-7167-1045-5. Edge dominating set (decision version) is discussed under the dominating set problem, which is the problem GT2 in Appendix A1.1. Minimum maximal matching (decision version) is the problem GT10 in Appendix A1.1.
- ^ Ausiello, Giorgio; Crescenzi, Pierluigi; Gambosi, Giorgio; Kann, Viggo; Marchetti-Spaccamela, Alberto; Protasi, Marco (2003), Complexity and Approximation: Combinatorial Optimization Problems and Their Approximability Properties, Springer. Minimum edge dominating set (optimisation version) is the problem GT3 in Appendix B (page 370). Minimum maximal matching (optimisation version) is the problem GT10 in Appendix B (page 374). See also Minimum Edge Dominating Set and Minimum Maximal Matching in the web compendium.
- ^ Leslie Valiant, The Complexity of Enumeration and Reliability Problems, SIAM J. Comput., 8(3), 410–421
- ^ Bezáková, Ivona; Štefankovič, Daniel; Vazirani, Vijay V.; Vigoda, Eric (2008). "Accelerating Simulated Annealing for the Permanent and Combinatorial Counting Problems". SIAM Journal on Computing. 37 (5): 1429–1454. doi:10.1137/050644033.
- ^ Callan, David (2009), A combinatorial survey of identities for the double factorial, arXiv:0906.1317.
- ^ Tichy, Robert F.; Wagner, Stephan (2005), "Extremal problems for topological indices in combinatorial chemistry" (PDF), Journal of Computational Biology, 12 (7): 1004–1013, doi:10.1089/cmb.2005.12.1004.
- ^ de Carvalho, Marcelo H.; Cheriyan, Joseph (2005), "An algorithm for ear decompositions of matching-covered graphs", Proc. ACM/SIAM Symposium on Discrete Algorithms (SODA), pp. 415–423.
- ^ Rabin, Michael O.; Vazirani, Vijay V. (1989), "Maximum matchings in general graphs through randomization", J. of Algorithms, 10: 557–567, doi:10.1016/0196-6774(89)90005-9.
- ^ Tassa, Tamir (2012), "Finding all maximally-matchable edges in a bipartite graph", Theoretical Computer Science, 423: 50–58, doi:10.1016/j.tcs.2011.12.071.
- ^ Gionis, Aris; Mazza, Arnon; Tassa, Tamir (2008), "k-Anonymization revisited", International Conference on Data Engineering (ICDE), pp. 744–753.
- ^ See, e.g., Trinajstić, Nenad; Klein, Douglas J.; Randić, Milan (1986), "On some solved and unsolved problems of chemical graph theory", International Journal of Quantum Chemistry, 30 (S20): 699–742, doi:10.1002/qua.560300762.
Further reading
- László Lovász; M. D. Plummer (1986), Matching Theory, North-Holland, ISBN 0-444-87916-1
- Thomas H. Cormen, Charles E. Leiserson, Ronald L. Rivest and Clifford Stein (2001), Introduction to Algorithms (second ed.), MIT Press and McGraw–Hill, Chapter 26, pp. 643–700, ISBN 0-262-53196-8
{{citation}}: CS1 maint: multiple names: authors list (link) - András Frank (2004). On Kuhn's Hungarian Method – A tribute from Hungary (PDF) (Technical report). Egerváry Research Group.
- Michael L. Fredman and Robert E. Tarjan (1987), "Fibonacci heaps and their uses in improved network optimization algorithms", Journal of the ACM, 34 (3): 595–615, doi:10.1145/28869.28874.
- S. J. Cyvin; Ivan Gutman (1988), Kekule Structures in Benzenoid Hydrocarbons, Springer-Verlag
{{citation}}: Unknown parameter|lastauthoramp=ignored (|name-list-style=suggested) (help) - Marek Karpinski and Wojciech Rytter (1998), Fast Parallel Algorithms for Graph Matching Problems, Oxford University Press, ISBN 978-0-19-850162-6