Enhancer (genetics)


- DNA
- Enhancer
- Promoter
- Gene
- Transcription Activator Protein
- Mediator Protein
- RNA Polymerase
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins (activators) to increase the likelihood that transcription of a particular gene will occur.[1][2] These proteins are usually referred to as transcription factors. Enhancers are cis-acting. They can be located up to 1 Mbp (1,000,000 bp) away from the gene, upstream or downstream from the start site.[2][3] There are hundreds of thousands of enhancers in the human genome.[2] They are found in both prokaryotes and eukaryotes.[4] Active enhancers typically get transcribed as enhancer or regulatory non-coding RNA, whose expression levels correlate with mRNA levels of target genes.[5][6]
The first discovery of a eukaryotic enhancer was in the immunoglobulin heavy chain gene in 1983.[7][8][9] This enhancer, located in the large intron, provided an explanation for the transcriptional activation of rearranged Vh gene promoters while unrearranged Vh promoters remained inactive.[10] Lately, enhancers have been shown to be involved in certain medical conditions, for example, myelosuppression.[11] Since 2022, scientists have used artificial intelligence to design synthetic enhancers and applied them in animal systems, first in a cell line,[12] and one year later also in vivo.[13][14]
Locations
[edit]In eukaryotic cells the structure of the chromatin complex of DNA is folded in a way that functionally mimics the supercoiled state characteristic of prokaryotic DNA, so although the enhancer DNA may be far from the gene in a linear way, it is spatially close to the promoter and gene. This allows it to interact with the general transcription factors and RNA polymerase II.[15] The same mechanism holds true for silencers in the eukaryotic genome. Silencers are antagonists of enhancers that, when bound to its proper transcription factors called repressors, repress the transcription of the gene. Silencers and enhancers may be in close proximity to each other or may even be in the same region only differentiated by the transcription factor the region binds to.
An enhancer may be located upstream or downstream of the gene it regulates. Furthermore, an enhancer does not need to be located near the transcription initiation site to affect transcription, as some have been found located several hundred thousand base pairs upstream or downstream of the start site.[16] Enhancers do not act on the promoter region itself, but are bound by activator proteins as first shown by in vivo competition experiments.[17][18] Subsequently, molecular studies showed direct interactions with transcription factors and cofactors, including the mediator complex, which recruits polymerase II and the general transcription factors which then begin transcribing the genes.[19][20] Enhancers can also be found within introns. An enhancer's orientation may even be reversed without affecting its function; additionally, an enhancer may be excised and inserted elsewhere in the chromosome, and still affect gene transcription.[9] That is one reason that introns polymorphisms may have effects although they are not translated.[citation needed] Enhancers can also be found at the exonic region of an unrelated gene[21][22][23] and they may act on genes on another chromosome.[24]
Enhancers are bound by p300-CBP and their location can be predicted by ChIP-seq against this family of coactivators.[25][26][27][28]
Role in gene expression
[edit]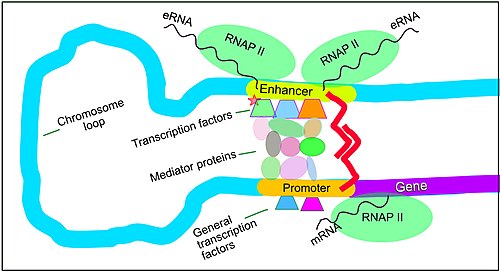
Gene expression in mammals is regulated by many cis-regulatory elements, including core promoters and promoter-proximal elements that are located near the transcription start sites of genes. Core promoters are sufficient to direct transcription initiation, but generally have low basal activity.[29] Other important cis-regulatory modules are localized in DNA regions that are distant from the transcription start sites. These include enhancers, silencers, insulators and tethering elements.[30] Among this constellation of elements, enhancers and their associated transcription factors have a leading role in the regulation of gene expression.[31] An enhancer localized in a DNA region distant from the promoter of a gene can have a very large effect on gene expression, with some genes undergoing up to 100-fold increased expression due to an activated enhancer.[32]
Enhancers are regions of the genome that are major gene-regulatory elements. Enhancers control cell-type-specific gene expression programs, most often by looping through long distances to come in physical proximity with the promoters of their target genes.[33] While there are hundreds of thousands of enhancer DNA regions,[2] for a particular type of tissue only specific enhancers are brought into proximity with the promoters that they regulate. In a study of brain cortical neurons, 24,937 loops were found, bringing enhancers to their target promoters.[32] Multiple enhancers, each often at tens or hundreds of thousands of nucleotides distant from their target genes, loop to their target gene promoters and can coordinate with each other to control the expression of their common target gene.[33]
The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene. The loop is stabilized by a dimer of a connector protein (e.g. dimer of CTCF or YY1), with one member of the dimer anchored to its binding motif on the enhancer and the other member anchored to its binding motif on the promoter (represented by the red zigzags in the illustration).[34] Several cell function specific transcription factors (there are about 1,600 transcription factors in a human cell[35]) generally bind to specific motifs on an enhancer[36] and a small combination of these enhancer-bound transcription factors, when brought close to a promoter by a DNA loop, govern level of transcription of the target gene. Mediator (a complex usually consisting of about 26 proteins in an interacting structure) communicates regulatory signals from enhancer DNA-bound transcription factors directly to the RNA polymerase II (pol II) enzyme bound to the promoter.[37]
Enhancers, when active, are generally transcribed from both strands of DNA with RNA polymerases acting in two different directions, producing two Enhancer RNAs (eRNAs) as illustrated in the Figure.[38] Like mRNAs, these eRNAs are usually protected by their 5′ cap.[39] An inactive enhancer may be bound by an inactive transcription factor. Phosphorylation of the transcription factor may activate it and that activated transcription factor may then activate the enhancer to which it is bound (see small red star representing phosphorylation of transcription factor bound to enhancer in the illustration).[40] An activated enhancer begins transcription of its RNA before activating transcription of messenger RNA from its target gene.[41]
Theories
[edit]As of 2005[update], there are two different theories on the information processing that occurs on enhancers:[42]
- Enhanceosomes – rely on highly cooperative, coordinated action and can be disabled by single point mutations that move or remove the binding sites of individual proteins.
- Flexible billboards – less integrative, multiple proteins independently regulate gene expression and their sum is read in by the basal transcriptional machinery.
Examples in the human genome
[edit]HACNS1
[edit]HACNS1 (also known as CENTG2 and located in the Human Accelerated Region 2) is a gene enhancer "that may have contributed to the evolution of the uniquely opposable human thumb, and possibly also modifications in the ankle or foot that allow humans to walk on two legs". Evidence to date shows that of the 110,000 gene enhancer sequences identified in the human genome, HACNS1 has undergone the most change during the evolution of humans following the split with the ancestors of chimpanzees.[citation needed]
GADD45G
[edit]An enhancer near the gene GADD45g has been described that may regulate brain growth in chimpanzees and other mammals, but not in humans.[43] The GADD45G regulator in mice and chimps is active in regions of the brain where cells that form the cortex, ventral forebrain, and thalamus are located and may suppress further neurogenesis. Loss of the GADD45G enhancer in humans may contribute to an increase of certain neuronal populations and to forebrain expansion in humans.[citation needed]
In developmental biology
[edit]The development, differentiation and growth of cells and tissues require precisely regulated patterns of gene expression. Enhancers work as cis-regulatory elements to mediate both spatial and temporal control of development by turning on transcription in specific cells and/or repressing it in other cells. Thus, the particular combination of transcription factors and other DNA-binding proteins in a developing tissue controls which genes will be expressed in that tissue. Enhancers allow the same gene to be used in diverse processes in space and time.[citation needed][44]
Identification and characterization
[edit]Traditionally, enhancers were identified by enhancer trap techniques using a reporter gene or by comparative sequence analysis and computational genomics. In genetically tractable models such as the fruit fly Drosophila melanogaster, for example, a reporter construct such as the lacZ gene can be randomly integrated into the genome using a P element transposon. If the reporter gene integrates near an enhancer, its expression will reflect the expression pattern driven by that enhancer. Thus, staining the flies for LacZ expression or activity and cloning the sequence surrounding the integration site allows the identification of the enhancer sequence.[45]
The development of genomic and epigenomic technologies, however, has dramatically changed the outlook for cis-regulatory modules (CRM) discovery. Next-generation sequencing (NGS) methods now enable high-throughput functional CRM discovery assays, and the vastly increasing amounts of available data, including large-scale libraries of transcription factor-binding site (TFBS) motifs, collections of annotated, validated CRMs, and extensive epigenetic data across many cell types, are making accurate computational CRM discovery an attainable goal. An example of NGS-based approach called DNase-seq have enabled identification of nucleosome-depleted, or open chromatin regions, which can contain CRM. More recently techniques such as ATAC-seq have been developed which require less starting material. Nucelosome depleted regions can be identified in vivo through expression of Dam methylase, allowing for greater control of cell-type specific enhancer identification.[46] Computational methods include comparative genomics, clustering of known or predicted TF-binding sites, and supervised machine-learning approaches trained on known CRMs. All of these methods have proven effective for CRM discovery, but each has its own considerations and limitations, and each is subject to a greater or lesser number of false-positive identifications.[47] In the comparative genomics approach, sequence conservation of non-coding regions can be indicative of enhancers. Sequences from multiple species are aligned, and conserved regions are identified computationally.[48] Identified sequences can then be attached to a reporter gene such as green fluorescent protein or lacZ to determine the in vivo pattern of gene expression produced by the enhancer when injected into an embryo. mRNA expression of the reporter can be visualized by in situ hybridization, which provides a more direct measure of enhancer activity, since it is not subjected to the complexities of translation and protein folding. Although much evidence has pointed to sequence conservation for critical developmental enhancers, other work has shown that the function of enhancers can be conserved with little or no primary sequence conservation. For example, the RET enhancers in humans have very little sequence conservation to those in zebrafish, yet both species' sequences produce nearly identical patterns of reporter gene expression in zebrafish.[48] Similarly, in highly diverged insects (separated by around 350 million years), similar gene expression patterns of several key genes was found to be regulated through similarly constituted CRMs although these CRMs do not show any appreciable sequence conservation detectable by standard sequence alignment methods such as BLAST.[49]
In segmentation of insects
[edit]The enhancers determining early segmentation in Drosophila melanogaster embryos are among the best characterized developmental enhancers. In the early fly embryo, the gap gene transcription factors are responsible for activating and repressing a number of segmentation genes, such as the pair rule genes. The gap genes are expressed in blocks along the anterior-posterior axis of the fly along with other maternal effect transcription factors, thus creating zones within which different combinations of transcription factors are expressed. The pair-rule genes are separated from one another by non-expressing cells. Moreover, the stripes of expression for different pair-rule genes are offset by a few cell diameters from one another. Thus, unique combinations of pair-rule gene expression create spatial domains along the anterior-posterior axis to set up each of the 14 individual segments. The 480 bp enhancer responsible for driving the sharp stripe two of the pair-rule gene even-skipped (eve) has been well-characterized. The enhancer contains 12 different binding sites for maternal and gap gene transcription factors. Activating and repressing sites overlap in sequence. Eve is only expressed in a narrow stripe of cells that contain high concentrations of the activators and low concentration of the repressors for this enhancer sequence. Other enhancer regions drive eve expression in 6 other stripes in the embryo.[50]
In vertebrate patterning
[edit]Establishing body axes is a critical step in animal development. During mouse embryonic development, Nodal, a transforming growth factor-beta superfamily ligand, is a key gene involved in patterning both the anterior-posterior axis and the left-right axis of the early embryo. The Nodal gene contains two enhancers: the Proximal Epiblast Enhancer (PEE) and the Asymmetric Enhancer (ASE). The PEE is upstream of the Nodal gene and drives Nodal expression in the portion of the primitive streak that will differentiate into the node (also referred to as the primitive node).[51] The PEE turns on Nodal expression in response to a combination of Wnt signaling plus a second, unknown signal; thus, a member of the LEF/TCF transcription factor family likely binds to a TCF binding site in the cells in the node. Diffusion of Nodal away from the node forms a gradient which then patterns the extending anterior-posterior axis of the embryo.[52] The ASE is an intronic enhancer bound by the fork head domain transcription factor Fox1. Early in development, Fox1-driven Nodal expression establishes the visceral endoderm. Later in development, Fox1 binding to the ASE drives Nodal expression on the left side of the lateral plate mesoderm, thus establishing left-right asymmetry necessary for asymmetric organ development in the mesoderm.[53]
Establishing three germ layers during gastrulation is another critical step in animal development. Each of the three germ layers has unique patterns of gene expression that promote their differentiation and development. The endoderm is specified early in development by Gata4 expression, and Gata4 goes on to direct gut morphogenesis later. Gata4 expression is controlled in the early embryo by an intronic enhancer that binds another forkhead domain transcription factor, FoxA2. Initially the enhancer drives broad gene expression throughout the embryo, but the expression quickly becomes restricted to the endoderm, suggesting that other repressors may be involved in its restriction. Late in development, the same enhancer restricts expression to the tissues that will become the stomach and pancreas. An additional enhancer is responsible for maintaining Gata4 expression in the endoderm during the intermediate stages of gut development.[54]
Multiple enhancers promote developmental robustness
[edit]Some genes involved in critical developmental processes contain multiple enhancers of overlapping function. Secondary enhancers, or "shadow enhancers", may be found many kilobases away from the primary enhancer ("primary" usually refers to the first enhancer discovered, which is often closer to the gene it regulates). On its own, each enhancer drives nearly identical patterns of gene expression. Are the two enhancers truly redundant? Recent work has shown that multiple enhancers allow fruit flies to survive environmental perturbations, such as an increase in temperature. When raised at an elevated temperature, a single enhancer sometimes fails to drive the complete pattern of expression, whereas the presence of both enhancers permits normal gene expression.[55]
Evolution of developmental mechanisms
[edit]One theme of research in evolutionary developmental biology ("evo-devo") is investigating the role of enhancers and other cis-regulatory elements in producing morphological changes via developmental differences between species.[citation needed]
Stickleback Pitx1
[edit]Recent work has investigated the role of enhancers in morphological changes in threespine stickleback fish. Sticklebacks exist in both marine and freshwater environments, but sticklebacks in many freshwater populations have completely lost their pelvic fins (appendages homologous to the posterior limb of tetrapods).
Pitx1 is a homeobox gene involved in posterior limb development in vertebrates. Preliminary genetic analyses indicated that changes in the expression of this gene were responsible for pelvic reduction in sticklebacks. Fish expressing only the freshwater allele of Pitx1 do not have pelvic spines, whereas fish expressing a marine allele retain pelvic spines. A more thorough characterization showed that a 500 base pair enhancer sequence is responsible for turning on Pitx1 expression in the posterior fin bud. This enhancer is located near a chromosomal fragile site—a sequence of DNA that is likely to be broken and thus more likely to be mutated as a result of imprecise DNA repair. This fragile site has caused repeated, independent losses of the enhancer responsible for driving Pitx1 expression in the pelvic spines in isolated freshwater population, and without this enhancer, freshwater fish fail to develop pelvic spines.[56]
In Drosophila wing pattern evolution
[edit]Pigmentation patterns provide one of the most striking and easily scored differences between different species of animals. Pigmentation of the Drosophila wing has proven to be a particularly amenable system for studying the development of complex pigmentation phenotypes. The Drosophila guttifera wing has 12 dark pigmentation spots and 4 lighter gray intervein patches. Pigment spots arise from expression of the yellow gene, whose product produces black melanin. Recent work has shown that two enhancers in the yellow gene produce gene expression in precisely this pattern – the vein spot enhancer drives reporter gene expression in the 12 spots, and the intervein shade enhancer drives reporter expression in the 4 distinct patches. These two enhancers are responsive to the Wnt signaling pathway, which is activated by wingless expression at all of the pigmented locations. Thus, in the evolution of the complex pigmentation phenotype, the yellow pigment gene evolved enhancers responsive to the wingless signal and wingless expression evolved at new locations to produce novel wing patterns.[57]
In inflammation and cancer
[edit]Each cell typically contains several hundred of a special class of enhancers that stretch over many kilobases long DNA sequences, called "super-enhancers".[58] These enhancers contain a large number of binding sites for sequence-specific, inducible transcription factors, and regulate expression of genes involved in cell differentiation.[59] During inflammation, the transcription factor NF-κB facilitates remodeling of chromatin in a manner that selectively redistributes cofactors from high-occupancy enhancers, thereby repressing genes involved in maintaining cellular identify whose expression they enhance; at the same time, this F-κB-driven remodeling and redistribution activates other enhancers that guide changes in cellular function through inflammation.[60][61] As a result, inflammation reprograms cells, altering their interactions with the rest of tissue and with the immune system.[62][63] In cancer, proteins that control NF-κB activity are dysregulated, permitting malignant cells to decrease their dependence on interactions with local tissue, and hindering their surveillance by the immune system.[64][65]
Designing enhancers in synthetic biology
[edit]Synthetic regulatory elements such as enhancers promise to be a powerful tool to direct gene products to particular cell types in order to treat disease by activating beneficial genes or by halting aberrant cell states.
Since 2022, artificial intelligence and transfer learning strategies have led to a better understanding of the features of regulatory DNA sequences, the prediction, and the design of synthetic enhancers.[66][67]
Building on work in cell culture,[66] synthetic enhancers were successfully applied to entire living organisms in 2023. Using deep neural networks, scientists simulated the evolution of DNA sequences to analyze the emergence of features that underly enhancer function. This allowed the design and production of a range of functioning synthetic enhancers for different cell types of the fruit fly brain.[14] A second approach trained artificial intelligence models on single-cell DNA accessibility data and transferred the learned models towards the prediction of enhancers for selected tissues in the fruit fly embryo. These enhancer prediction models were used to design synthetic enhancers for the nervous system, brain, muscle, epidermis and gut.[13]
See also
[edit]References
[edit]- ^ Blackwood EM, Kadonaga JT (July 1998). "Going the distance: a current view of enhancer action". Science. 281 (5373): 60–63. Bibcode:1998Sci...281...60.. doi:10.1126/science.281.5373.60. PMID 9679020. S2CID 11666739.
- ^ a b c d Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G (April 2013). "Enhancers: five essential questions". Nature Reviews. Genetics. 14 (4): 288–295. doi:10.1038/nrg3458. PMC 4445073. PMID 23503198.
- ^ Maston GA, Evans SK, Green MR (2006). "Transcriptional regulatory elements in the human genome". Annual Review of Genomics and Human Genetics. 7: 29–59. doi:10.1146/annurev.genom.7.080505.115623. PMID 16719718. S2CID 12346247.
- ^ Kulaeva OI, Nizovtseva EV, Polikanov YS, Ulianov SV, Studitsky VM (December 2012). "Distant activation of transcription: mechanisms of enhancer action". Molecular and Cellular Biology. 32 (24): 4892–4897. doi:10.1128/MCB.01127-12. PMC 3510544. PMID 23045397.
- ^ Yosefzon Y, David C, Tsukerman A, Pnueli L, Qiao S, Boehm U, et al. (19 September 2017). "An epigenetic switch repressing Tet1 in gonadotropes activates the reproductive axis". Proceedings of the National Academy of Sciences. 114 (38): 10131–10136. doi:10.1073/pnas.1704393114. ISSN 0027-8424. PMC 5617270. PMID 28855337.
- ^ Burren OS, Rubio García A, Javierre BM, Rainbow DB, Cairns J, Cooper NJ, et al. (4 September 2017). "Chromosome contacts in activated T cells identify autoimmune disease candidate genes". Genome Biology. 18 (1): 165. doi:10.1186/s13059-017-1285-0. ISSN 1474-760X. PMC 5584004. PMID 28870212.
- ^ Mercola M, Wang XF, Olsen J, Calame K (August 1983). "Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus". Science. 221 (4611): 663–665. Bibcode:1983Sci...221..663M. doi:10.1126/science.6306772. PMID 6306772.
- ^ Banerji J, Olson L, Schaffner W (July 1983). "A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes". Cell. 33 (3): 729–740. doi:10.1016/0092-8674(83)90015-6. PMID 6409418. S2CID 23981549.
- ^ a b Gillies SD, Morrison SL, Oi VT, Tonegawa S (July 1983). "A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene". Cell. 33 (3): 717–728. doi:10.1016/0092-8674(83)90014-4. PMID 6409417. S2CID 40313833.
- ^ Hauptman G, Reichert MC, Abdal Rhida MA, Evans TA (December 2022). "Characterization of enhancer fragments in Drosophila robo2". Fly. 16 (1): 312–346. bioRxiv 10.1101/2022.08.01.502399. doi:10.1080/19336934.2022.2126259. PMC 9559326. PMID 36217698.
- ^ Zhigulev A, Norberg Z, Cordier J, Spalinskas R, Bassereh H, Björn N, et al. (March 2024). "Enhancer mutations modulate the severity of chemotherapy-induced myelosuppression". Life Science Alliance. 7 (3): e202302244. doi:10.26508/lsa.202302244. PMC 10796589. PMID 38228368.
- ^ de Almeida BP, Reiter F, Pagani M, Stark A (May 2022). "DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers". Nature Genetics. 54 (5): 613–624. doi:10.1038/s41588-022-01048-5. PMID 35551305.
- ^ a b de Almeida BP, Schaub C, Pagani M, Secchia S, Furlong EE, Stark A (February 2024). "Targeted design of synthetic enhancers for selected tissues in the Drosophila embryo". Nature. 626 (7997): 207–211. Bibcode:2024Natur.626..207D. doi:10.1038/s41586-023-06905-9. PMC 10830412. PMID 38086418.
- ^ a b Taskiran II, Spanier KI, Dickmänken H, Kempynck N, Pančíková A, Ekşi EC, et al. (February 2024). "Cell-type-directed design of synthetic enhancers". Nature. 626 (7997): 212–220. Bibcode:2024Natur.626..212T. doi:10.1038/s41586-023-06936-2. PMC 10830415. PMID 38086419.
- ^ Maston GA, Evans SK, Green MR (1 January 2006). "Transcriptional regulatory elements in the human genome". Annual Review of Genomics and Human Genetics. 7 (1): 29–59. doi:10.1146/annurev.genom.7.080505.115623. PMID 16719718. S2CID 12346247.
- ^ Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gómez-Marín C, et al. (March 2014). "Obesity-associated variants within FTO form long-range functional connections with IRX3". Nature. 507 (7492): 371–375. Bibcode:2014Natur.507..371S. doi:10.1038/nature13138. PMC 4113484. PMID 24646999.
- ^ Mercola M, Goverman J, Mirell C, Calame K (January 1985). "Immunoglobulin heavy-chain enhancer requires one or more tissue-specific factors". Science. 227 (4684): 266–270. Bibcode:1985Sci...227..266M. doi:10.1126/science.3917575. PMID 3917575.
- ^ Scholer H, Haslinger A, Heguy A, Holtgreve H, Karin M (April 1986). "In vivo competition between a metallothionein regulatory element and the SV40 enhancer". Science. 232 (4746): 76–80. Bibcode:1986Sci...232...76S. doi:10.1126/science.3006253. PMID 3006253.
- ^ Kuras L, Borggrefe T, Kornberg RD (November 2003). "Association of the Mediator complex with enhancers of active genes". Proceedings of the National Academy of Sciences of the United States of America. 100 (24): 13887–13891. Bibcode:2003PNAS..10013887K. doi:10.1073/pnas.2036346100. PMC 283516. PMID 14623974.
- ^ Ramasamy S, Aljahani A, Karpinska MA, Cao TB, Velychko T, Cruz JN, et al. (July 2023). "The Mediator complex regulates enhancer-promoter interactions". Nature Structural & Molecular Biology. 30 (7): 991–1000. doi:10.1038/s41594-023-01027-2. PMC 10352134. PMID 37430065.
- ^ Dong X, Navratilova P, Fredman D, Drivenes Ø, Becker TS, Lenhard B (March 2010). "Exonic remnants of whole-genome duplication reveal cis-regulatory function of coding exons". Nucleic Acids Research. 38 (4): 1071–1085. doi:10.1093/nar/gkp1124. PMC 2831330. PMID 19969543.
- ^ Birnbaum RY, Clowney EJ, Agamy O, Kim MJ, Zhao J, Yamanaka T, et al. (June 2012). "Coding exons function as tissue-specific enhancers of nearby genes". Genome Research. 22 (6): 1059–1068. doi:10.1101/gr.133546.111. PMC 3371700. PMID 22442009.
- ^ Eichenlaub MP, Ettwiller L (November 2011). "De novo genesis of enhancers in vertebrates". PLOS Biology. 9 (11): e1001188. doi:10.1371/journal.pbio.1001188. PMC 3206014. PMID 22069375.
- ^ Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA (June 2005). "Interchromosomal associations between alternatively expressed loci". Nature. 435 (7042): 637–645. Bibcode:2005Natur.435..637S. doi:10.1038/nature03574. PMID 15880101. S2CID 1755326.
- ^ Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, et al. (September 2009). "Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes". Cell. 138 (5): 1019–1031. doi:10.1016/j.cell.2009.06.049. PMC 2750862. PMID 19698979.
- ^ Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. (May 2009). "Histone modifications at human enhancers reflect global cell-type-specific gene expression". Nature. 459 (7243): 108–112. Bibcode:2009Natur.459..108H. doi:10.1038/nature07829. PMC 2910248. PMID 19295514.
- ^ Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. (February 2009). "ChIP-seq accurately predicts tissue-specific activity of enhancers". Nature. 457 (7231): 854–858. Bibcode:2009Natur.457..854V. doi:10.1038/nature07730. PMC 2745234. PMID 19212405.
- ^ Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. (September 2010). "ChIP-Seq identification of weakly conserved heart enhancers". Nature Genetics. 42 (9): 806–810. doi:10.1038/ng.650. PMC 3138496. PMID 20729851.
- ^ Haberle V, Stark A (October 2018). "Eukaryotic core promoters and the functional basis of transcription initiation". Nature Reviews. Molecular Cell Biology. 19 (10): 621–637. doi:10.1038/s41580-018-0028-8. PMC 6205604. PMID 29946135.
- ^ Verheul TC, van Hijfte L, Perenthaler E, Barakat TS (2020). "The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1". Frontiers in Cell and Developmental Biology. 8: 592164. doi:10.3389/fcell.2020.592164. PMC 7554316. PMID 33102493.
- ^ Spitz F, Furlong EE (September 2012). "Transcription factors: from enhancer binding to developmental control". Nature Reviews. Genetics. 13 (9): 613–626. doi:10.1038/nrg3207. PMID 22868264. S2CID 205485256.
- ^ a b Beagan JA, Pastuzyn ED, Fernandez LR, Guo MH, Feng K, Titus KR, et al. (June 2020). "Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression". Nature Neuroscience. 23 (6): 707–717. doi:10.1038/s41593-020-0634-6. PMC 7558717. PMID 32451484.
- ^ a b Schoenfelder S, Fraser P (August 2019). "Long-range enhancer-promoter contacts in gene expression control". Nature Reviews. Genetics. 20 (8): 437–455. doi:10.1038/s41576-019-0128-0. PMID 31086298. S2CID 152283312.
- ^ Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, et al. (December 2017). "YY1 Is a Structural Regulator of Enhancer-Promoter Loops". Cell. 171 (7): 1573–1588.e28. doi:10.1016/j.cell.2017.11.008. PMC 5785279. PMID 29224777.
- ^ Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. (February 2018). "The Human Transcription Factors". Cell. 172 (4): 650–665. doi:10.1016/j.cell.2018.01.029. PMID 29425488.
- ^ Grossman SR, Engreitz J, Ray JP, Nguyen TH, Hacohen N, Lander ES (July 2018). "Positional specificity of different transcription factor classes within enhancers". Proceedings of the National Academy of Sciences of the United States of America. 115 (30): E7222–E7230. Bibcode:2018PNAS..115E7222G. doi:10.1073/pnas.1804663115. PMC 6065035. PMID 29987030.
- ^ Allen BL, Taatjes DJ (March 2015). "The Mediator complex: a central integrator of transcription". Nature Reviews. Molecular Cell Biology. 16 (3): 155–166. doi:10.1038/nrm3951. PMC 4963239. PMID 25693131.
- ^ Mikhaylichenko O, Bondarenko V, Harnett D, Schor IE, Males M, Viales RR, et al. (January 2018). "The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription". Genes & Development. 32 (1): 42–57. doi:10.1101/gad.308619.117. PMC 5828394. PMID 29378788.
- ^ Yao L, Liang J, Ozer A, Leung AK, Lis JT, Yu H (July 2022). "A comparison of experimental assays and analytical methods for genome-wide identification of active enhancers". Nature Biotechnology. 40 (7): 1056–1065. doi:10.1038/s41587-022-01211-7. PMC 9288987. PMID 35177836.
- ^ Li QJ, Yang SH, Maeda Y, Sladek FM, Sharrocks AD, Martins-Green M (January 2003). "MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300". The EMBO Journal. 22 (2): 281–291. doi:10.1093/emboj/cdg028. PMC 140103. PMID 12514134.
- ^ Carullo NV, Phillips Iii RA, Simon RC, Soto SA, Hinds JE, Salisbury AJ, et al. (September 2020). "Enhancer RNAs predict enhancer-gene regulatory links and are critical for enhancer function in neuronal systems". Nucleic Acids Research. 48 (17): 9550–9570. doi:10.1093/nar/gkaa671. PMC 7515708. PMID 32810208.
- ^ Arnosti DN, Kulkarni MM (April 2005). "Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards?" (PDF). Journal of Cellular Biochemistry. 94 (5): 890–898. doi:10.1002/jcb.20352. PMID 15696541. S2CID 32405464. Archived from the original (PDF) on 21 July 2006. Retrieved 8 August 2019.
- ^ McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, et al. (March 2011). "Human-specific loss of regulatory DNA and the evolution of human-specific traits". Nature. 471 (7337): 216–219. Bibcode:2011Natur.471..216M. doi:10.1038/nature09774. PMC 3071156. PMID 21390129.
- ^ Barrett LW, Fletcher S, Wilton SD (November 2012). "Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements". Cellular and Molecular Life Sciences. 69 (21): 3613–3634. doi:10.1007/s00018-012-0990-9. PMC 3474909. PMID 22538991.
- ^ Hartenstein V, Jan YN (June 1992). "Studying Drosophila embryogenesis with P-lacZ enhancer trap lines". Roux's Archives of Developmental Biology. 201 (4): 194–220. doi:10.1007/BF00188752. PMID 28305845. S2CID 25759655.
- ^ Aughey GN, Estacio Gomez A, Thomson J, Yin H, Southall TD (February 2018). "CATaDa reveals global remodelling of chromatin accessibility during stem cell differentiation in vivo". eLife. 7. doi:10.7554/eLife.32341. PMC 5826290. PMID 29481322.
- ^ Suryamohan K, Halfon MS (2014). "Identifying transcriptional cis-regulatory modules in animal genomes". Wiley Interdisciplinary Reviews. Developmental Biology. 4 (2): 59–84. doi:10.1002/wdev.168. PMC 4339228. PMID 25704908.
- ^ a b Visel A, Bristow J, Pennacchio LA (February 2007). "Enhancer identification through comparative genomics". Seminars in Cell & Developmental Biology. 18 (1): 140–152. doi:10.1016/j.semcdb.2006.12.014. PMC 1855162. PMID 17276707.
- ^ "Evidence for Deep Regulatory Similarities in Early Developmental Programs across Highly Diverged Insects". Genome Biology and Evolution. Archived from the original on 10 July 2015.
- ^ Borok MJ, Tran DA, Ho MC, Drewell RA (January 2010). "Dissecting the regulatory switches of development: lessons from enhancer evolution in Drosophila". Development. 137 (1): 5–13. doi:10.1242/dev.036160. PMC 2796927. PMID 20023155.
- ^ Norris DP, Robertson EJ (June 1999). "Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements". Genes & Development. 13 (12): 1575–1588. doi:10.1101/gad.13.12.1575. PMC 316799. PMID 10385626.
- ^ Granier C, Gurchenkov V, Perea-Gomez A, Camus A, Ott S, Papanayotou C, et al. (January 2011). "Nodal cis-regulatory elements reveal epiblast and primitive endoderm heterogeneity in the peri-implantation mouse embryo". Developmental Biology. 349 (2): 350–362. doi:10.1016/j.ydbio.2010.10.036. PMID 21047506.
- ^ Norris DP, Brennan J, Bikoff EK, Robertson EJ (July 2002). "The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo". Development. 129 (14): 3455–3468. doi:10.1242/dev.129.14.3455. PMID 12091315. S2CID 24426844.
- ^ Rojas A, Schachterle W, Xu SM, Martín F, Black BL (October 2010). "Direct transcriptional regulation of Gata4 during early endoderm specification is controlled by FoxA2 binding to an intronic enhancer". Developmental Biology. 346 (2): 346–355. doi:10.1016/j.ydbio.2010.07.032. PMC 2945415. PMID 20692247.
- ^ Perry MW, Boettiger AN, Bothma JP, Levine M (September 2010). "Shadow enhancers foster robustness of Drosophila gastrulation". Current Biology. 20 (17): 1562–1567. Bibcode:2010CBio...20.1562P. doi:10.1016/j.cub.2010.07.043. PMC 4257487. PMID 20797865.
- ^ Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, et al. (January 2010). "Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer". Science. 327 (5963): 302–305. Bibcode:2010Sci...327..302C. doi:10.1126/science.1182213. PMC 3109066. PMID 20007865.
- ^ Werner T, Koshikawa S, Williams TM, Carroll SB (April 2010). "Generation of a novel wing colour pattern by the Wingless morphogen". Nature. 464 (7292): 1143–1148. Bibcode:2010Natur.464.1143W. doi:10.1038/nature08896. PMID 20376004. S2CID 4407744.
- ^ Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. (April 2013). "Master transcription factors and mediator establish super-enhancers at key cell identity genes". Cell. 153 (2): 307–319. doi:10.1016/j.cell.2013.03.035. PMC 3653129. PMID 23582322.
- ^ Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, et al. (October 2013). "Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants". Proceedings of the National Academy of Sciences of the United States of America. 110 (44): 17921–17926. Bibcode:2013PNAS..11017921P. doi:10.1073/pnas.1317023110. PMC 3816444. PMID 24127591.
- ^ Brown JD, Lin CY, Duan Q, Griffin G, Federation A, Paranal RM, et al. (October 2014). "NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis". Molecular Cell. 56 (2): 219–231. doi:10.1016/j.molcel.2014.08.024. PMC 4224636. PMID 25263595.
- ^ Schmidt SF, Larsen BD, Loft A, Nielsen R, Madsen JG, Mandrup S (September 2015). "Acute TNF-induced repression of cell identity genes is mediated by NFκB-directed redistribution of cofactors from super-enhancers". Genome Research. 25 (9): 1281–1294. doi:10.1101/gr.188300.114. PMC 4561488. PMID 26113076.
- ^ Chatterjee B, Banoth B, Mukherjee T, Taye N, Vijayaragavan B, Chattopadhyay S, et al. (December 2016). "Late-phase synthesis of IκBα insulates the TLR4-activated canonical NF-κB pathway from noncanonical NF-κB signaling in macrophages". Science Signaling. 9 (457): ra120. doi:10.1126/scisignal.aaf1129. PMC 5260935. PMID 27923915.
- ^ Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SC, Erdos MR, et al. (April 2015). "Super-enhancers delineate disease-associated regulatory nodes in T cells". Nature. 520 (7548): 558–562. Bibcode:2015Natur.520..558V. doi:10.1038/nature14154. PMC 4409450. PMID 25686607.
- ^ Vlahopoulos SA, Cen O, Hengen N, Agan J, Moschovi M, Critselis E, et al. (August 2015). "Dynamic aberrant NF-κB spurs tumorigenesis: a new model encompassing the microenvironment". Cytokine & Growth Factor Reviews. 26 (4): 389–403. doi:10.1016/j.cytogfr.2015.06.001. PMC 4526340. PMID 26119834.
- ^ Zou Z, Huang B, Wu X, Zhang H, Qi J, Bradner J, et al. (May 2014). "Brd4 maintains constitutively active NF-κB in cancer cells by binding to acetylated RelA". Oncogene. 33 (18): 2395–2404. doi:10.1038/onc.2013.179. PMC 3913736. PMID 23686307.
- ^ a b de Almeida BP, Reiter F, Pagani M, Stark A (May 2022). "DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers". Nature Genetics. 54 (5): 613–624. doi:10.1038/s41588-022-01048-5. PMID 35551305.
- ^ Avsec Ž, Weilert M, Shrikumar A, Krueger S, Alexandari A, Dalal K, et al. (March 2021). "Base-resolution models of transcription-factor binding reveal soft motif syntax". Nature Genetics. 53 (3): 354–366. doi:10.1038/s41588-021-00782-6. PMC 8812996. PMID 33603233.
External links
[edit]- Enhancer+Elements,Genetic at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- TFSEARCH
- JASPAR
- ReMap
- ENCODE threads explorer Enhancer discovery and characterization. Nature
