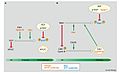
SCF complex
Skp, Cullin, F-box containing complex (or SCF complex) is a multi-protein E3 ubiquitin ligase complex that catalyzes the ubiquitination of proteins destined for 26S proteasomal degradation.[1] Along with the anaphase-promoting complex,[2] SCF has important roles in the ubiquitination of proteins involved in the cell cycle. The SCF complex also marks various other cellular proteins for destruction.[3]
Core components
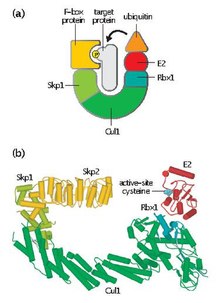
[edit]SCF contains a variable F-box protein and three core subunits:
- F-box protein (FBP) – FBP contributes to the substrate specificity of the SCF complex by first aggregating to target proteins independently of the complex. Each FBP (e.g. Skp2) may recognize several different substrates in a manner that is dependent on post-translational modifications such as phosphorylation or glycosylation. FBP then binds to Skp1 of the SCF complex using an F-box motif, bringing the target protein into proximity with the functional E2 ubiquitin-conjugating enzyme. FBP is also essential in regulating SCF activity during the course of the cell cycle. SCF levels are thought to remain constant throughout the cell-cycle. Instead, FBP affinity for protein substrates is regulated through cyclin-CDK-mediated phosphorylation of target proteins.[4]
- Skp1 – Skp1 is an adaptor protein that is essential for the recognition and binding of F-box proteins.
- Cullin (CUL1) – Cullin forms the major structural scaffold of the SCF complex and links the skp1 domain to the Rbx1 domain. Different combinations of Cullin and FBPs can generate on the order of a hundred types of E3 ubiquitin ligases that target different substrates.[5]
- RBX1 – Rbx1 contains a small, zinc-binding Really Interesting New Gene (RING) finger domain, to which the E2 ubiquitin-conjugating enzyme binds. This binding event allows the transferral of ubiquitin from E2 to a lysine residue on the target protein.
Discovery
[edit]The first hint that led to the discovery of the SCF complex came from genetic screens of Saccharomyces cerevisiae, also known as budding yeast. Temperature-sensitive cell division cycle (Cdc) mutants—such as Cdc4, Cdc34, and Cdc53[6]—arrested in G1 with unreplicated DNA and multiple elongated buds.[7] The phenotype was attributed to a failure to degrade Sic1, an inhibitor of S cyclin-CDK complexes.[6] These findings indicated that proteolysis is important in the G1/S transition.
Next, biochemical studies revealed that Cdc34 is an E2 enzyme that physically interacts with an E3 ubiquitin ligase complex containing Skp1, Cdc4, and several other proteins.[6] Skp1’s known binding partners—specifically Skp2, Cyclin F, and Cdc4—were found to share an approximately 40 residue motif that was coined the F-box motif. The F-box hypothesis[8] that followed these discoveries proposed that F-box proteins recruit substrates targeted for degradation, and that Skp1 links the F-box protein to the core ubiquitination complex.
Subsequent genetic studies in Caenorhabditis elegans later contributed to the elucidation of other SCF complex components.[8]
Cell cycle regulation
[edit]The eukaryotic cell cycle[9] is regulated through the synthesis, degradation, binding interactions, post-translational modifications of regulatory proteins. Of these regulatory proteins, two ubiquitin ligases are crucial for progression through cell cycle checkpoints. The anaphase-promoting complex (APC) controls the metaphase-anaphase transition, while the SCF complex controls G1/S and G2/M transitions. Specifically, SCF has been shown to regulate centriole splitting from late telophase to the G1/S transition.[1]
SCF activity is largely regulated by post-translational modifications. For instance, ubiquitin-mediated autocatalytic degradation of FBPs is a mechanism of decreasing SCF activity.
Well-characterized cell cycle substrates of SCF complexes include:
- cyclin family proteins: Cyclin D, Cyclin E[2]
- transcriptional regulators: Myc, E2f1, p130[2]
- cyclin-dependent kinase inhibitors (CKIs): p27Kip1, p21, Wee1[2]
- centriole proteins: Cep250, Ninein[1]
There are approximately seventy human FBPs, several of which are involved in cell cycle control as a component of SCF complexes.[10]
Skp2 is an FBP that binds CKIs such as p27Kip1 and p21.[11] Skp2 binds p27Kip1 only when two conditions are met: p27Kip1 is phosphorylated by E/A/CKD2 and bound to Cks1. As a consequence of binding Skp2, p27Kip1 is ubiquitinated and targeted for degradation in late G1 and early S.[4] SCF-Skp2 also targets p130 for degradation in a phosphorylation dependent manner.
Beta-transducin repeat-containing protein (βTRCP) is an FBP that targets emi1—an APC/C-Cdh1 inhibitor—and wee1 for degradation during early mitosis.[2] βTRCP recognizes these substrates after they are phosphorylated by Polo-like kinase 1 or Cyclin B-CDK1.
Fbw7, which is the human homolog of cdc4 in yeast, is an FBP that targets Cyclin E, Myc, Notch and c-Jun for degradation.[4] Fbw7 is stable throughout the cell cycle[12] and is localized to the nucleus due to the presence of a nuclear localization sequence (NLS).[13] SCF-Fbw7 targets Sic1—when at least six out of nine possible sites are phosphorylated—and Swi5 for degradation.[14] Since Sic1 normally prevents premature entry into S-phase by inhibiting Cyclin B-CDK1, targeting Sic1 for degradation promotes S-phase entry. Fbw7 is known to be a haplo-insufficient tumor suppressor gene implicated in several sporadic carcinomas, for which one mutant allele is enough to disturb the wild type phenotype.[15]
Fbxo4 is another tumor suppressor FBP that has been implicated in human carcinomas. SCF-fbxo4 plays a role in cell cycle control by targeting cyclin D1 for degradation.[4]
Cyclin F is an FBP that is associated with amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).[16][17] Mutations that prevent phosphorylation of Cyclin F alter the activity of SCF-Cyclin F, which likely affects downstream processes pertinent to neuron degeneration in ALS and FTD.[17] Normally, Cyclin F targets E2f1 for degradation.
Cancer
[edit]Recently, SCF complexes have become an attractive anti-cancer target because of their upregulation in some human cancers and their biochemically distinct active sites.[18] Though many of the aforementioned FBPs have been implicated in cancer, cytotoxicity has been a limiting factor of drug development.[19]
Skp2-targeting anti-sense oligonucleotides and siRNAs are in the drug development pipeline. Preliminary studies have shown that Skp2 downregulation can inhibit the growth of melanomas, lung cancer cells, oral cancer cells, and glioblastoma cells.[19]
βTRCP-targeting siRNAs have been shown to sensitize breast cancer cells and cervical cancer cells to existing chemotherapies.[19]
Plant hormone signaling
[edit]The plant hormone auxin binds Tir1 (Transport Inhibitor Response 1). Tir1 is an Auxin Signaling F-box Protein (AFB) that acts as an auxin receptor. Auxin-bound Tir1 stimulates binding of SCF-Tir1 to the AUX/IAA repressor. Subsequent degradation of the repressor results in activation of AUX/IAA (i.e. auxin-responsive) genes.[20]
The plant hormone Jasmonate binds Coi1, an FBP. SCF-Coi1 then binds the JAZ transcription factor and targets it for degradation. Degradation of the JAZ transcription factor allows for the transcription of the jasmonate responsive genes.[21]
References
[edit]- ^ a b c Ou, Young; Rattner, J.B. (2004), "The Centrosome in Higher Organisms: Structure, Composition, and Duplication", International Review of Cytology, 238, Elsevier: 119–182, doi:10.1016/s0074-7696(04)38003-4, ISBN 978-0-12-364642-2, PMID 15364198
- ^ a b c d e Fischer, Martin; Dang, Chi V.; DeCaprio, James A. (2018), "Control of Cell Division", Hematology, Elsevier, pp. 176–185, doi:10.1016/b978-0-323-35762-3.00017-2, ISBN 978-0-323-35762-3
- ^ Morgan, David "Protein Degradation in Cell-Cycle Control", The Cell Cycle; Principles of Control 2007
- ^ a b c d The Molecular Basis of Cancer. Elsevier. 2008. doi:10.1016/b978-1-4160-3703-3.x5001-7. ISBN 978-1-4160-3703-3.
- ^ Hegde, Ashok N. (2010), "Ubiquitin-Dependent Protein Degradation", Comprehensive Natural Products II, Elsevier, pp. 699–752, doi:10.1016/b978-008045382-8.00697-3, ISBN 978-0-08-045382-8, retrieved 2019-12-01
- ^ a b c Patton, E (1998-06-01). "Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis". Trends in Genetics. 14 (6): 236–243. doi:10.1016/s0168-9525(98)01473-5. ISSN 0168-9525. PMID 9635407.
- ^ Schwob, E (1994-10-21). "The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae". Cell. 79 (2): 233–244. doi:10.1016/0092-8674(94)90193-7. ISSN 0092-8674. PMID 7954792. S2CID 34939988.
- ^ a b Willems, Andrew R.; Schwab, Michael; Tyers, Mike (November 2004). "A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1695 (1–3): 133–170. doi:10.1016/j.bbamcr.2004.09.027. ISSN 0167-4889. PMID 15571813.
- ^ Vodermaier, Hartmut C. (September 2004). "APC/C and SCF: Controlling Each Other and the Cell Cycle". Current Biology. 14 (18): R787 – R796. doi:10.1016/j.cub.2004.09.020. ISSN 0960-9822. PMID 15380093.
- ^ Lambrus, Bramwell G.; Moyer, Tyler C.; Holland, Andrew J. (2017-08-31). "Applying the auxin-inducible degradation (AID) system for rapid protein depletion in mammalian cells". doi:10.1101/182840.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Frescas, David; Pagano, Michele (June 2008). "Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer". Nature Reviews Cancer. 8 (6): 438–449. doi:10.1038/nrc2396. ISSN 1474-175X. PMC 2711846. PMID 18500245.
- ^ Mathias, Neal; Steussy, C. Nic; Goebl, Mark G. (1998-02-13). "An Essential Domain within Cdc34p Is Required for Binding to a Complex Containing Cdc4p and Cdc53p inSaccharomyces cerevisiae". Journal of Biological Chemistry. 273 (7): 4040–4045. doi:10.1074/jbc.273.7.4040. ISSN 0021-9258. PMID 9461595.
- ^ Blondel, M. (2000-11-15). "Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4". The EMBO Journal. 19 (22): 6085–6097. doi:10.1093/emboj/19.22.6085. ISSN 1460-2075. PMC 305831. PMID 11080155.
- ^ Kishi, T.; Ikeda, A.; Koyama, N.; Fukada, J.; Nagao, R. (2008-09-11). "A refined two-hybrid system reveals that SCFCdc4-dependent degradation of Swi5 contributes to the regulatory mechanism of S-phase entry". Proceedings of the National Academy of Sciences. 105 (38): 14497–14502. Bibcode:2008PNAS..10514497K. doi:10.1073/pnas.0806253105. ISSN 0027-8424. PMC 2567208. PMID 18787112.
- ^ Calhoun, Eric S.; Jones, Jessa B.; Ashfaq, Raheela; Adsay, Volkan; Baker, Suzanne J.; Valentine, Virginia; Hempen, Paula M.; Hilgers, Werner; Yeo, Charles J.; Hruban, Ralph H.; Kern, Scott E. (October 2003). "BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) Mutations in Distinct Subsets of Pancreatic Cancer". The American Journal of Pathology. 163 (4): 1255–1260. doi:10.1016/s0002-9440(10)63485-2. ISSN 0002-9440. PMC 1868306. PMID 14507635.
- ^ Yu, Yujiao; Nakagawa, Tadashi; Morohoshi, Akane; Nakagawa, Makiko; Ishida, Noriko; Suzuki, Naoki; Aoki, Masashi; Nakayama, Keiko (2019-09-30). "Pathogenic mutations in the ALS gene CCNF cause cytoplasmic mislocalization of Cyclin F and elevated VCP ATPase activity". Human Molecular Genetics. 28 (20): 3486–3497. doi:10.1093/hmg/ddz119. ISSN 0964-6906. PMID 31577344.
- ^ a b Lee, Albert; Rayner, Stephanie L.; De Luca, Alana; Gwee, Serene S. L.; Morsch, Marco; Sundaramoorthy, Vinod; Shahheydari, Hamideh; Ragagnin, Audrey; Shi, Bingyang; Yang, Shu; Williams, Kelly L. (October 2017). "Casein kinase II phosphorylation of cyclin F at serine 621 regulates the Lys48-ubiquitylation E3 ligase activity of the SCF (cyclin F) complex". Open Biology. 7 (10): 170058. doi:10.1098/rsob.170058. ISSN 2046-2441. PMC 5666078. PMID 29021214.
- ^ Skaar, Jeffrey R.; Pagan, Julia K.; Pagano, Michele (December 2014). "SCF ubiquitin ligase-targeted therapies". Nature Reviews Drug Discovery. 13 (12): 889–903. doi:10.1038/nrd4432. ISSN 1474-1776. PMC 4410837. PMID 25394868.
- ^ a b c Sun, L. Jia and Y. (2011-02-28). "SCF E3 Ubiquitin Ligases as Anticancer Targets". Current Cancer Drug Targets. 11 (3): 347–356. doi:10.2174/156800911794519734. PMC 3323109. PMID 21247385.
- ^ Dharmasiri, Nihal; Dharmasiri, Sunethra; Estelle, Mark (May 2005). "The F-box protein TIR1 is an auxin receptor". Nature. 435 (7041): 441–445. Bibcode:2005Natur.435..441D. doi:10.1038/nature03543. ISSN 0028-0836. PMID 15917797. S2CID 4428049.
- ^ Devoto, Alessandra; Nieto-Rostro, Manuela; Xie, Daoxin; Ellis, Christine; Harmston, Rebecca; Patrick, Elaine; Davis, Jackie; Sherratt, Leigh; Coleman, Mark; Turner, John G. (November 2002). "COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex inArabidopsis". The Plant Journal. 32 (4): 457–466. doi:10.1046/j.1365-313x.2002.01432.x. ISSN 0960-7412. PMID 12445118.
-
Morgan, David "Protein Degradation in Cell-Cycle Control", The Cell Cycle; Principles of Control 2007
-
Hartmut C. Vodermaier (2004). "APC/C and SCF: Controlling Each Other and the Cell Cycle", Current Biology, 14 (787)