MUSCLE (alignment software)
| Original author(s) | Robert C. Edgar |
|---|---|
| Developer(s) | drive5 |
| Initial release | 2004 |
| Stable release | 3.8.31
/ 18 August 2016 |
| Repository | github |
| Operating system | Linux, macOS, Windows |
| Platform | IA-32, x86-64 |
| Available in | English |
| Type | Multiple sequence alignment |
| License | Public domain |
| Website | drive5 |
MUltiple Sequence Comparison by Log-Expectation (MUSCLE) is a computer software for multiple sequence alignment of protein and nucleotide sequences. It is licensed as public domain. The method was published by Robert C. Edgar in two papers in 2004. The first paper, published in Nucleic Acids Research, introduced the sequence alignment algorithm.[1] The second paper, published in BMC Bioinformatics, presented more technical details.[2]
Algorithm
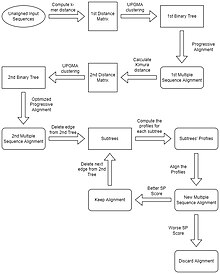
[edit]The MUSCLE algorithm proceeds in three stages: the draft progressive, improved progressive, and refinement stage.
Stage 1: Draft Progressive
[edit]In this first stage, the algorithm produces a multiple alignment, emphasizing speed over accuracy. This step begins by computing the k-mer distance for every pair of input sequences to create a distance matrix. UPGMA clusters the distance matrix to produce a binary tree. From this tree a progressive alignment is constructed, beginning with the creation of profiles for each leaf of the tree. For every node in the tree, a pairwise alignment is constructed of the two child profiles, creating a new profile to be assigned to that node. This continues until there is a multiple sequence alignment of all input sequences at the root of the tree.[1]
Stage 2: Improved Progressive
[edit]This stage focuses on obtaining a more optimal tree by calculating the Kimura distance for each pair of input sequences using the multiple sequence alignment obtained in Stage one, and creates a second distance matrix. UPGMA clusters this distance matrix to obtain a second binary tree. A progressive alignment is performed to obtain a multiple sequence alignment like in Stage 1, but it is optimized by only computing alignments in subtrees whose branching orders have changed from the first binary tree, resulting in a more accurate alignment.[1]
Stage 3: Refinement
[edit]In this final stage, an edge is chosen from the second tree, with edges being visited in decreasing distance from the root. The chosen edge is deleted, dividing the tree into two subtrees. The profile of the multiple alignment is then computed for each subtree. A new multiple sequence alignment is produced by re-aligning the subtree profiles. If the SP score is improved, the new alignment is kept, otherwise, it is discarded. The process of deleting an edge and aligning is repeated until convergence, or until a user-defined limit is reached.[1]
Complexity and Comparison
[edit]In the first two stages of the algorithm, the time complexity is O(N2L + NL2), the space complexity is O(N2 + NL + L2). The refinement stage adds to the time complexity another term, O(N3L).[1] MUSCLE is often used as a replacement for Clustal, since it usually (but not always) gives better sequence alignments, depending on the chosen options. is significantly faster than Clustal, more so for larger alignments.[1][2]
Algorithm Flowchart
[edit]Integration
[edit]MUSCLE is integrated into DNASTAR's Lasergene software, Geneious, and MacVector and is available in Sequencher, MEGA, and UGENE as a plug-in. MUSCLE is also available as a web service via the European Molecular Biology Laboratory (EMBL)-European Bioinformatics Institute (EBI).[3] As of September 2016, the two papers describing MUSCLE have been cited more than 19,000 times in total.[4]
See also
[edit]References
[edit]- ^ a b c d e f Edgar RC (2004). "MUSCLE: multiple sequence alignment with high accuracy and high throughput". Nucleic Acids Research. 32 (5): 1792–97. doi:10.1093/nar/gkh340. PMC 390337. PMID 15034147.
- ^ a b Edgar RC (2004). "MUSCLE: a multiple sequence alignment method with reduced time and space complexity". BMC Bioinformatics. 5 (1): 113. doi:10.1186/1471-2105-5-113. PMC 517706. PMID 15318951.
- ^ "MUSCLE < Multiple Sequence Alignment < EMBL-EBI". Archived from the original on 18 January 2015. Retrieved 1 September 2014.
- ^ "Robert C. Edgar - Google Scholar Citations". Archived from the original on 24 September 2016. Retrieved 1 September 2016.