Peptoid
This article needs additional citations for verification. (May 2011) |
Peptoids (root from the Greek πεπτός, peptós "digested"; derived from πέσσειν, péssein "to digest" and the Greek-derived suffix -oid meaning "like, like that of, thing like a ______," ), or poly-N-substituted glycines, are a class of biochemicals known as biomimetics that replicate the behavior of biological molecules.[1] Peptidomimetics are recognizable by side chains that are appended to the nitrogen atom of the peptide backbone, rather than to the α-carbons (as they are in amino acids).
Chemical structure and synthesis
[edit]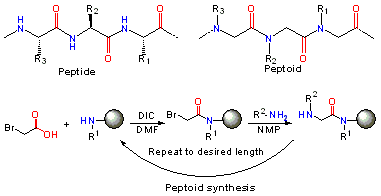
In peptoids, the side chain is connected to the nitrogen of the peptide backbone, instead of the α-carbon as in peptides. Notably, peptoids lack the amide hydrogen which is responsible for many of the secondary structure elements in peptides and proteins. Peptoids were first invented by Reyna J. Simon, Ronald N. Zuckermann, Paul Bartlett and Daniel V. Santi to mimic protein/peptide products to aid in the discovery of protease-stable small molecule drugs for the East Bay company Chiron.[2][3]
Following the sub-monomer protocol originally created by Ron Zuckermann,[4] each residue is installed in two steps: acylation and displacement. In the acylation step, a haloacetic acid, typically bromoacetic acid activated by diisopropylcarbodiimide reacts with the amine of the previous residue. In the displacement step (a classical SN2 reaction), an amine displaces the halide to form the N-substituted glycine residue. The submonomer approach allows the use of any commercially available or synthetically accessible amine with great potential for combinatorial chemistry.
Unique characteristics
[edit]Like D-Peptides and β peptides, peptoids are completely resistant to proteolysis,[5] and are therefore advantageous for therapeutic applications where proteolysis is a major issue. Since secondary structure in peptoids does not involve hydrogen bonding, it is not typically denatured by solvent, temperature, or chemical denaturants such as urea (see details below).
Notably, since the amino portion of the amino acid results from the use of any amine, thousands of commercially available amines can be used to generate unprecedented chemical diversity at each position at costs far lower than would be required for similar peptides or peptidomimetics. To date, at least 230 different amines have been used as side chains in peptoids.[6]
Structure
[edit]Peptoid oligomers are known to be conformationally unstable, due to the flexibility of the main-chain methylene groups and the absence of stabilizing hydrogen bond interactions along the backbone. Nevertheless, through the choice of appropriate side chains it is possible to form specific steric or electronic interactions that favour the formation of stable secondary structures like helices,[7] especially peptoids with C-α-branched side chains are known to adopt structure analogous to polyproline I helix.[8] Different strategies have been employed to predict and characterize peptoid secondary structure, with the ultimate goal of developing fully folded peptoid protein structures[9] The cis/trans amide bond isomerization still leads to a conformational heterogeneity which doesn’t allow for the formation of homogeneous peptoid foldamers.[10] Nonetheless, scientists were able to find trans-inducer N-Aryl side chains promoting polyproline type II helix,[11] and strong cis-inducer such as bulky naphtylethyl[12] and tert-butyl[13] side chains. It was also found that n→π* interactions can modulate the ratio of cis/trans amide bond conformers,[14] until reaching a complete control of the cis conformer in the peptoid backbone using a functionalizable triazolium side chain.[15]
Applications
[edit]The first demonstration of the use of peptoids was in screening a combinatorial library of diverse peptoids, which yielded novel high-affinity ligands for 7-transmembrane G-protein-couple receptors.[16]
Peptoids have been developed as candidates for a range of different biomedical applications,[17][18] including antimicrobial agents,[19] synthetic lung surfactants,[20][21] ligands for various proteins including Src Homology 3 (SH3 domain),[22] Vascular Endothelial Growth Factor (VEGF) receptor 2,[23] and antibody Immunoglobulin G biomarkers for the identification of Alzheimer's disease.[24]
Due to their advantageous characteristics as described above, peptoids are also being actively developed for use in nanotechnology,[25] an area in which they may play an important role.[26]
Antimicrobial agents
[edit]Researchers supported by grants from the NIH and NIAID tested the efficacy of antimicrobial peptoids against antibiotic-resistant strands of Mycobacterium tuberculosis.[27] Antimicrobial peptoids demonstrate a non-specific mechanism of action against the bacterial membrane, one that differs from small-molecule antibiotics that bind to specific receptors (and thus are susceptible to mutations or alterations in bacterial structure). Preliminary results suggested "appreciable activity" against drug-sensitive bacterial strands, leading to a call for more research into the viability of peptoids as a new class of tuberculocidal drugs.[27]
Researchers at the Barron Lab at Stanford University (supported by a NIH Pioneer Award grant) are currently studying whether upregulation of the human host defense peptide LL-37 or application of antimicrobial treatments based on LL-37 may prevent or treat sporadic Alzheimer’s dementia. Lead researcher Annelise Barron discovered that the innate human defense peptide LL-37 binds to the peptide Ab, which is associated with Alzheimer's disease. Barron's insight is that an imbalance between LL-37 and Ab may be a critical factor affecting AD-associated fibrils and plaques. The project extends focus upon the potential relationship between chronic, oral P. gingivalis and herpesvirus (HSV-1) infections to the progression of Alzheimer's dementia.
See also
[edit]References
[edit]- ^ "What if We Could Give Viruses a One-Two Punch? by Berkeley Lab on Exposure". Exposure. Retrieved 2021-09-16.
- ^ Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, et al. (October 1992). "Peptoids: a modular approach to drug discovery". Proceedings of the National Academy of Sciences of the United States of America. 89 (20): 9367–9371. doi:10.1073/pnas.89.20.9367. PMC 50132. PMID 1409642.
- ^ "What if We Could Give Viruses a One-Two Punch? by Berkeley Lab on Exposure". Exposure. Retrieved 2021-09-17.
- ^ Zuckermann RN, Kerr JM, Kent SB, Moos WH (1992). "Efficient method for the preparation of peptoids [oligo(N-substituted glycines)] by submonomer solid-phase synthesis". Journal of the American Chemical Society. 114 (26): 10646–10647. doi:10.1021/ja00052a076.
- ^ Miller SM, Simon RJ, Ng S, Zuckermann RN, Kerr JM, Moos WH (May 1995). "Comparison of the proteolytic susceptibilities of homologous L‐amino acid, D‐amino acid, and N‐substituted glycine peptide and peptoid oligomers". Drug Development Research. 35 (1): 20–32. doi:10.1002/ddr.430350105. S2CID 84619520.
- ^ Culf AS, Ouellette RJ (August 2010). "Solid-phase synthesis of N-substituted glycine oligomers (alpha-peptoids) and derivatives". Molecules. 15 (8): 5282–5335. doi:10.3390/molecules15085282. PMC 6257730. PMID 20714299.
- ^ Kirshenbaum K, Barron AE, Goldsmith RA, Armand P, Bradley EK, Truong KT, et al. (April 1998). "Sequence-specific polypeptoids: a diverse family of heteropolymers with stable secondary structure". Proceedings of the National Academy of Sciences of the United States of America. 95 (8): 4303–4308. doi:10.1073/pnas.95.8.4303. PMC 22484. PMID 9539732.
- ^ Armand P, Kirshenbaum K, Goldsmith RA, Farr-Jones S, Barron AE, Truong KT, et al. (April 1998). "NMR determination of the major solution conformation of a peptoid pentamer with chiral side chains". Proceedings of the National Academy of Sciences of the United States of America. 95 (8): 4309–4314. doi:10.1073/pnas.95.8.4309. PMC 22485. PMID 9539733.
- ^ Wetzler M, Barron AE (2011). "Progress in the de novo design of structured peptoid protein mimics". Biopolymers. 96 (5): 556–560. doi:10.1002/bip.21621. PMID 22180903.
- ^ Yoo B, Kirshenbaum K (December 2008). "Peptoid architectures: elaboration, actuation, and application". Current Opinion in Chemical Biology. 12 (6): 714–721. doi:10.1016/j.cbpa.2008.08.015. PMID 18786652.
- ^ Shah NH, Butterfoss GL, Nguyen K, Yoo B, Bonneau R, Rabenstein DL, Kirshenbaum K (December 2008). "Oligo(N-aryl glycines): a new twist on structured peptoids". Journal of the American Chemical Society. 130 (49): 16622–16632. doi:10.1021/ja804580n. PMID 19049458.
- ^ Stringer JR, Crapster JA, Guzei IA, Blackwell HE (October 2011). "Extraordinarily robust polyproline type I peptoid helices generated via the incorporation of α-chiral aromatic N-1-naphthylethyl side chains". Journal of the American Chemical Society. 133 (39): 15559–15567. doi:10.1021/ja204755p. PMC 3186054. PMID 21861531.
- ^ Roy O, Caumes C, Esvan Y, Didierjean C, Faure S, Taillefumier C (May 2013). "The tert-butyl side chain: a powerful means to lock peptoid amide bonds in the cis conformation". Organic Letters. 15 (9): 2246–2249. doi:10.1021/ol400820y. PMID 23590358.
- ^ Gorske BC, Stringer JR, Bastian BL, Fowler SA, Blackwell HE (November 2009). "New strategies for the design of folded peptoids revealed by a survey of noncovalent interactions in model systems". Journal of the American Chemical Society. 131 (45): 16555–16567. doi:10.1021/ja907184g. PMC 3175426. PMID 19860427.
- ^ Caumes C, Roy O, Faure S, Taillefumier C (June 2012). "The click triazolium peptoid side chain: a strong cis-amide inducer enabling chemical diversity". Journal of the American Chemical Society. 134 (23): 9553–9556. doi:10.1021/ja302342h. PMID 22612307.
- ^ Zuckermann RN, Martin EJ, Spellmeyer DC, Stauber GB, Shoemaker KR, Kerr JM, et al. (August 1994). "Discovery of nanomolar ligands for 7-transmembrane G-protein-coupled receptors from a diverse N-(substituted)glycine peptoid library". Journal of Medicinal Chemistry. 37 (17): 2678–2685. doi:10.1021/jm00043a007. PMID 8064796.
- ^ Fowler SA, Blackwell HE (April 2009). "Structure-function relationships in peptoids: recent advances toward deciphering the structural requirements for biological function". Organic & Biomolecular Chemistry. 7 (8): 1508–1524. doi:10.1039/b817980h. PMC 5962266. PMID 19343235.
- ^ Zuckermann RN, Kodadek T (June 2009). "Peptoids as potential therapeutics". Current Opinion in Molecular Therapeutics. 11 (3): 299–307. PMID 19479663.
- ^ Diamond G, Molchanova N, Herlan C, Fortkort JA, Lin JS, Figgins E, et al. (March 2021). "Potent Antiviral Activity against HSV-1 and SARS-CoV-2 by Antimicrobial Peptoids". Pharmaceuticals. 14 (4): 304. doi:10.3390/ph14040304. PMC 8066833. PMID 33807248.
- ^ Brown NJ, Johansson J, Barron AE (October 2008). "Biomimicry of surfactant protein C". Accounts of Chemical Research. 41 (10): 1409–1417. doi:10.1021/ar800058t. PMC 3270935. PMID 18834153.
- ^ Czyzewski AM, McCaig LM, Dohm MT, Broering LA, Yao LJ, Brown NJ, et al. (May 2018). "Effective in vivo treatment of acute lung injury with helical, amphipathic peptoid mimics of pulmonary surfactant proteins". Scientific Reports. 8 (1): 6795. doi:10.1038/s41598-018-25009-3. PMC 5931611. PMID 29717157.
- ^ Nguyen JT, Turck CW, Cohen FE, Zuckermann RN, Lim WA (December 1998). "Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors". Science. 282 (5396): 2088–2092. doi:10.1126/science.282.5396.2088. PMID 9851931.
- ^ Udugamasooriya DG, Dineen SP, Brekken RA, Kodadek T (April 2008). "A peptoid "antibody surrogate" that antagonizes VEGF receptor 2 activity". Journal of the American Chemical Society. 130 (17): 5744–5752. doi:10.1021/ja711193x. PMID 18386897.
- ^ Reddy MM, Wilson R, Wilson J, Connell S, Gocke A, Hynan L, et al. (January 2011). "Identification of candidate IgG biomarkers for Alzheimer's disease via combinatorial library screening". Cell. 144 (1): 132–142. doi:10.1016/j.cell.2010.11.054. PMC 3066439. PMID 21215375.
- ^ Nam KT, Shelby SA, Choi PH, Marciel AB, Chen R, Tan L, et al. (May 2010). "Free-floating ultrathin two-dimensional crystals from sequence-specific peptoid polymers". Nature Materials. 9 (5): 454–460. doi:10.1038/nmat2742. PMID 20383129.
- ^ Drexler KE (2011). "Peptoids at the 7th Summit: toward macromolecular systems engineering". Biopolymers. 96 (5): 537–544. doi:10.1002/bip.21623. PMID 22180902. S2CID 36456597.
- ^ a b Kapoor R, Eimerman PR, Hardy JW, Cirillo JD, Contag CH, Barron AE (June 2011). "Efficacy of antimicrobial peptoids against Mycobacterium tuberculosis". Antimicrobial Agents and Chemotherapy. 55 (6): 3058–3062. doi:10.1128/AAC.01667-10. PMC 3101442. PMID 21464254.
