Wood-decay fungus



A wood-decay or xylophagous fungus is any species of fungus that digests moist wood, causing it to rot. Some species of wood-decay fungi attack dead wood, such as brown rot, and some, such as Armillaria (honey fungus), are parasitic and colonize living trees. Excessive moisture above the fibre saturation point in wood is required for fungal colonization and proliferation.[1] In nature, this process causes the breakdown of complex molecules and leads to the return of nutrients to the soil.[2] Wood-decay fungi consume wood in various ways; for example, some attack the carbohydrates in wood, and some others decay lignin. The rate of decay of wooden materials in various climates can be estimated by empirical models.[3]
Wood-decay fungi can be classified according to the type of decay that they cause. The best-known types are brown rot, soft rot, and white rot.[4][5] Each produce different enzymes, can degrade different plant materials, and can colonise different environmental niches.[6] Brown rot and soft rot both digest a tree's cellulose and hemicellulose but not its lignin; white rot digests lignin as well. The residual products of decomposition from fungal action have variable pH, solubility and redox potentials. Over time this residue becomes incorporated in the soil and sediment so can have a noticeable effect on the environment of that area.[6]
Wood decay fungi are considered key species in the forest ecosystems [7] because the process of decomposing dead wood creates new habitats for other species, helps in the nutrient recycling, participate in the energy transportation and transformation and provides food to other species.[8] They are also used as indicator species for conservation projects.
Wood decay fungi are dependent on wood. Due to forestry, cutting trees and removal of decaying wood, many species are classified as threatened.[9][10]
Brown rot
[edit]
Brown-rot fungi break down hemicellulose and cellulose that form the wood structure. Cellulose is broken down by hydrogen peroxide (H2O2) that is produced during the breakdown of hemicellulose.[4] Because hydrogen peroxide is a small molecule, it can diffuse rapidly through the wood, leading to a decay that is not confined to the direct surroundings of the fungal hyphae. As a result of this type of decay, the wood shrinks, shows a brown discoloration, and cracks into roughly cubical pieces, a phenomenon termed cubical fracture. The fungi of certain types remove cellulose compounds from wood, and hence the wood turns brown.[citation needed]
Brown rot in a dry, crumbly condition is sometimes incorrectly referred to as dry rot in general. The term brown rot replaced the general use of the term dry rot, as wood must be damp to decay, although it may become dry later. Dry rot is a generic name for certain species of brown-rot fungi. Brown-rot fungi of particular economic importance include Serpula lacrymans (true dry rot), Fibroporia vaillantii (mine fungus), and Coniophora puteana (cellar fungus), which may attack timber in buildings. Other brown-rot fungi include the sulfur shelf, Phaeolus schweinitzii, and Fomitopsis pinicola.[11]
Brown-rot fungal decay is characterised by extensive demethylation of lignins whereas white-rot tends to produce low yields of molecules with demethylated functional groups.[12] There are very few brown rot fungi in tropical climates or in southern temperate zones. Most brown rot fungi have a geographical range north of the Tropic of Cancer (23.5° latitude), and most of these are found north of the 35° latitude, corresponding to a roughly boreal distribution. Those brown rot fungi between latitudes 23.5° and 35° are typically found at high elevations in pine forest regions, or in coniferous forest regions such as the Rocky Mountains or the Himalayas.[13]
Soft rot
[edit]
Soft-rot fungi secrete cellulase from their hyphae, an enzyme that breaks down cellulose in wood.[4] This leads to the formation of microscopic cavities inside the wood and, sometimes, to a discoloration and cracking-pattern, similar to brown rot.[4][5] Soft-rot fungi need fixed nitrogen in order to synthesize enzymes, which they obtain either from the wood or from the environment. Examples of soft-rot-causing fungi are Chaetomium, Ceratocystis, and Kretzschmaria deusta.[citation needed]
Soft-rot fungi are able to colonise conditions that are normally too hot, cold or wet for brown- or white-rot to inhabit. They can also decompose woods containing high levels of protective from the compounds that are resistant to biological attack; the bark of many woody plants contains a high concentration of tannins, which are difficult for fungi to decompose, as well as suberin, which may act as a microbial barrier.[14] The bark acts as a form of protection for the more vulnerable interior of the plant.[14] Soft-rot fungi are, apparently, not able to decompose matter as effectively as white-rot fungi, as they are less aggressive decomposers.[6]
White rot
[edit]
White-rot fungi are a type of fungi comprising agaricomycetes, basidiomycetes, and some ascomycetes that are capable of decomposing many tree species. It is now recognized that saprotrophic interactions have profound effects on forest biomes.[15] White-rot fungi are characterized by their ability to break down the lignin, cellulose, and hemicellulose of wood. As a result of this ability, white-rot fungi are considered a vital component of the carbon cycle, because of their ability to access carbon pools that would otherwise remain inaccessible. The name “white rot” derives from the white color and rotting texture of the remaining crystalline cellulose from wood degraded by these fungi.[16] Most knowledge of white-rot fungi comes from Coriolus versicolor and Phanerochaete chrysosporium.[17] White-rot fungi show strong participation in interspecific competition, culminating in the evolution of lignin catabolism specificity. The current and future applications of white-rot fungi as a potential component of mycoremediation merit greater study of these saprotrophs.[18]
Biochemistry
[edit]Compared to other saprotrophs, white-rot fungi possess the specialized ability to cleave lignin into smaller, more processable molecules. Lignin is a biopolymer which combines with cellulose to form the lignocellulose complex, an important complex that confers strength and durability to plant cell walls. Lignin is a macromolecule formed from the combination of many phenolic aromatic groups via oxidative coupling. Because of its high stability, lignin is incapable of being broken down through simple decomposition. As a result, white-rot fungi employ a series of enzymes that break lignin down into smaller aromatic rings. The relative abundance of phenylpropane alkyl side chains of lignin characteristically decreases when decayed by white-rot fungi.[12] Since lignin is the specialized food source of white-rot fungi, understanding the two different catabolic pathways is important.
Lignin metabolism through peroxidases
[edit]The first way white-rot fungi can break down lignin involves a high-redox-potential catalyzed peroxidase attack on the heme pocket, thus reducing the stability of lignin. The process starts with creation of extracellular hydrogen peroxide (H2O2), a process completed via glyoxal oxidase (GLX). Extracellular hydrogen peroxide may be responsible for creation of hydroxyl radical (·OH) via the Fenton reaction: Fe2+ + H2O2 → Fe3+ + ·OH + OH−[19] The peroxidases used to oxidize lignin are lignin peroxidase (LiP), manganese peroxidase (MnP), and versatile peroxidase (VP).[20] These peroxidases are commonly referred to as fungal class II peroxidases (PODs). Research suggests there may be another group of POD enzymes: basal peroxidases, including novel peroxidase (NoP). The NoP of Postia placenta is characterized by its inability to bind Mn2+ and its low redox potential.[21] PODs developed in the common ancestor of white-rot, brown-rot and mycorrhizal fungi but these enzyme families have undergone secondary loss or contraction in the latter two groups.[22] LiPs are oxidioreductases specific to lignin degradation. VPs are a class of peroxidase that combines elements of both LiPs and MnPs. LiPs and VPs are specific to heme product architecture allowing direct oxidation of benzene groups regardless of linkages.[23] Direct oxidation of benzene groups results in the creation of an unstable radical aromatic. However, the hydrogen peroxide, bound to the heme group on the heme pocket, is unable to access the bulky lignin due to steric hindrance. As a result, LiP and VP enzymes create a tryptophan radical on their protein surface which allows long-range electron transfer from the aromatic substrate to the activated cofactor.[24]
Lignin metabolism through laccase
[edit]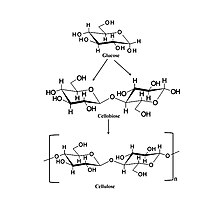
The second mechanism for breaking down lignin involves laccase, a low-redox-potential oxidase incapable of direct attack. Laccase can be used both in breaking and forming lignin. It cleaves lignin by reducing oxygen, creating a free radical which allows a hydroxyl radical (·OH) to attack the ring and deposit an alcohol group (OH). Deprotonation follows, resulting in the breaking of C-C (aryl-alphaC) bond into two aromatic rings. These products enter the fungal hyphae to be further broken down via catabolic processes. After the lignin complex is broken down, other saprotrophs can enter and begin degrading the newly created products.[16] The final products of these transformations are carbon dioxide and water. While it is known that brown-rot fungi can also target lignin, they are only capable of modifying and are not capable of completely recycling it with a few exceptions.[19] The ability to degrade lignin, previously supposed to only occur in white-rot fungi which have PODs, was found in Botryobasidium botryosum and Jappia argillacea, two brown-rot fungi, lacking PODs. While the general pathway is currently unknown, research supports the existence of a continuum of features that separate the two fungal types rather than distinct categories.[25]
Cellulose metabolism
[edit]While white-rot fungi specialized in catabolizing lignin, they are also capable of metabolizing other common organic forms of carbon like cellulose. Cellulose is also a laborious molecule to cleave.[26] First, cellobiohydrolases, found in all white-rot fungi, hydrolyze the 1,4-beta-D-glycosidic bonds partially degrading cellulose.[27] GH61 enzymes initiate a copper-dependent oxidative (LPMO) attack on crystalline cellulose. LPMOs boost degradation by activating oxygen using a copper-containing histidine brace that increases glycoside hydrolase activity, effectively lowering the activation cost of the reaction, making cleavage much cheaper, and therefore, more profitable for the fungi.[26] Products from the cleavage are glucose and cellobiose. Another method involves endoglucanases hydrolyzing cellulose at random points before cellobiohydrolases cleave the chains, resulting in cellobiose. At the end of both processes, Beta-glucosidases further catabolize cellobiose into glucose.[16]
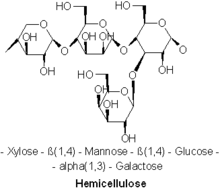
Hemicellulose metabolism
[edit]Another main food source of white-rot fungi is hemicellulose, a heteropolymer like cellulose that is not exclusively catabolized by white-rot fungi. The prevalent hemicellulose found in soft wood trees is Galactoglucomannan, a molecule made up of b-1,4-linked D-mannopyranose and D-glucopyranose units. Endo-1,4-b-D-mannanase breaks the prior linkages along the main chain of galactoglucomannan.[28] Recent studies have found that LPMOs, previously only thought to be used in cellulose cleavage, were also found to be important in the catabolism of hemicellulose in conjunction with glycoside hydrolase enzymes (GHs).[29] The availability of non white-rot fungi to catabolize cellulose and hemicellulose results in the creation of interspecific competition for access to these resources. Understanding the methods white-rot fungi use to dominate a resource and prevent competition will prove an important facet to understanding white-rot fungi.[citation needed]
Ecology
[edit]White rot competitive ability
[edit]Since white-rot fungi aren't the only saprotrophs capable of accessing cellulose and hemicellulose, competition ensues. Researchers attempted to estimate the effect of competition on white rot fungi. They reported that in sterile environments with no microbiota competitors present, white-rot fungi had good growth, but in soil with natural microbiota present, white-rot growth was variable. Even though white-rot fungi have a very specialized process for acquiring carbon, they are still vulnerable to competitors. Researchers clarified that white-rot fungi survival is dependent on its ability to defend lignocellulose substrate against attack by soil microbiota and its ability to establish itself within the soil bulk. These findings suggest that white-rot fungi and soil microbiota remain largely antagonistic in interactions, with only the highly competitive Pleurotus species capable of establishing themselves with only negligible negative impact due to soil microbiota. Less competitive white-rot fungi either failed to establish or produced lower enzyme concentrations associated with respiration. Successful interactions are characterized by which microbe arrives first and establishes a foothold.[30]
Brown-rot fungi and white-rot fungi have similar interspecific mycelial interactions. When white-rot fungal species occupied the same host distinct districts formed known as ‘decay columns’. Interactions were classified as interspecific competition.[31] There were two important results when competition occurs: ‘deadlock’, when neither species could dominate the other; and ‘replacement’ when once species achieved complete colonization and replaced the other. A different study noted a third option: ‘reciprocal replacement’ when fungi successfully captured some territory and simultaneously lost other territory.[32] Mutualism between two white-rot fungi was noted to be very rare.[31]
Findings suggested the important distinction between primary competition, that is competition to colonize unoccupied territory and antagonistic capture and defense of territory. Many competitive interactions were ‘intransitive’, meaning interactions involved more than two fungal species each often deploying a different antagonistic mechanism that gave it an advantage over one species but a disadvantage over others. Research further highlighted the importance of environmental factors including temperature, water potential, and invertebrate interactions in influencing competition. Findings suggested that competition increased decay, due to competition being expensive and saprotrophs needing to access more resources to fund it. Similarly, decay rates increased in smaller environments where natural resources were limited and competition intense. Even though brown-rot fungi lack the ability to decompose lignin, a relatively energetically expensive molecule, brown-rot fungi were slightly more competitive than white-rot fungi since they could still access the relatively cheaper cellulose and hemicellulose and devote more energy to competition and less to extracting nutrients.[32] Further evidence for white-rot fungi possessing long-term advantages was found in a study that determined that a longer time was required for white-rot fungal invasion of wood chips than for foliage litters. The data they collected on white-rot mass loss was sigmoid-shaped. This finding suggests that while white-rot fungi are not as competitive at decomposing carbon from common sources as other decomposers within the first year, but they proved to be more competitive after one year due to their specialized ability to access carbon from lignin.[33]
Competition is not just limited between fungi. The presence of white rot fungi, in this case Hypholoma fasciculare and Resinicium bicolor, on sterilized beech wood blocks resulted in a lower number of wood-inhabiting bacteria, even though lignin is not a food source of these bacteria.[34] This finding points to an antagonistic relationship between white-rot fungi and bacteria that both compete for cellulose and hemicellulose, as well as the existence of bactericidal and bacteriostatic weapons utilized by white-rot fungi against competitor bacteria. Though the mechanism is unknown, researchers suggested that white-rot fungi may utilize lignin decomposing enzymes, hydroxyl radicals, and aryl alcohols to create a toxic environment. Further environmental manipulation involved the release of PODs to lower the pH and create a more acidic habitat.[citation needed] The resulting conclusion is that peroxides not only make lignin accessible, but create a more accessible environment for white-rot fungi to compete in. Even with a specialized catabolic mechanism, competition remains a highly selective force on white-rot evolution.
Evolution
[edit]Insight on the evolutionary development of white-rot fungi comes from the evolution of lignin catabolism. Lignin is a precursor to the development of coal. During the Carboniferous (360-300 mya) and Permian (300-250 mya) there was a very high carbon accumulation. However, near the end of the Permian there was a sharp decline in carbon accumulation. White-rot fungi and their ability to cleave lignin evolved at the end of the Permian period.[35] Researchers attempted to reconstruct the evolution of saprotrophic capabilities. Results suggested that white-rot saprotrophs were the common ancestors of brown-rot fungi and ectomycorrhiza (ECM), but that in the latter two groups genes coding for PODs were lost.[36]
To gain insight on the evolution of lignolytic peroxidases, researchers resurrected ancestral lignolytic peroxidases from the Polyporales, a basidiomycete order that emerged 150 mya, and analyzed the lineage from that ancestor to the modern P. chrysosporium. One of the major findings was that ancestral versatile peroxidase (AVP) was not capable of functioning efficiently at low pH, a characteristic associated with modern LiPs. Findings also suggested that AVP possessed a much wider substrate specificity, the loss of which being an evolutionary cost of developing further specificity.[37]
Early peroxidases were unable to directly degrade lignin and relied on metal cations to separate phenol groups. Only later would peroxidases acquire the ability use a tryptophanyl radical, interacting with a bulky polymer at the surface of the peroxidase, to attack non-phenolic lignin. These findings highlight the importance of taking plant evolution into account when analyzing the evolution of white-rot fungus. Researchers note that plant cell walls have been steadily increasing and show evidence of convergent evolution. White-rot PODs also demonstrated convergent evolution. As plant cell walls have become more efficient, so have the peroxidases that destroy them.[38]
Researchers attempted to further understand the evolutionary development of white-rot fungi by using bioinformatics. They analyzed sixty-two genomes of Agaricomycetes of white-rot, brown-rot, ECM and other nutritional modes. Given that both white-rot and brown-rot share the ability to cleave cellulose and hemicellulose, they suggest that PODs developed after cellulolytic enzymes and that white-rot mechanisms were an elaboration based on the already existing saprotrophic model, not just on the utilization of PODs.[39] Understanding the evolutionary development of white-rot fungi provides insight onto a variety of potential uses.[citation needed]
Current and future applications
[edit]White-rot fungi have historically been valued as food, but in recent years exploration of their enzymatic capabilities has revealed white-rot fungi’s potential in depollution. White-rot fungi have long since been staples of human diet and remain an important source of nutrition for people around the world. White-rot fungi are commercially grown as a source of food – for example the shiitake mushroom, which in 2003 constituted approximately 25% of total mushroom production.[40] Due to white-rot fungi’s important ability to degrade lignin, they have been increasingly explored as potential sources in mycoremediation applications, applications focused on removing organic pollutants from the environment. All three enzyme types of lignin decomposition (LiPs, MnP, and Laccase) have been explored. White-rot fungi have been determined to degrade chlorinated aromatic hydrocarbons (CAHs), DDT, lindane, polycyclic aromatic hydrocarbons, polychlorinated biphenyls, PCP, polychlorinated dibenzo(p)dioxins, and azo dyes when studied in Phanerochaete chrysosporium, Trametes versicolor, Bjerkandere adusta, and Pleurotus ostreatus.[30] Noted limitations of white-rot fungi as pollutant cleaners is due to difficulty establishing the fungi in non-natural conditions. Other applications include biosorption, a process where biomass is utilized to remove solute wastes preventing pollution. Researchers studied the effect white-rot fungi could have on absorbing heavy metal ions via alginic acid, a linear polysaccharide composed of 1,4-linked beta-D-mannuronic and alpha-L-guluronic acid. The findings from the study indicated that Fungalia trogii was capable of biosorption of Hg2+, Cd2+, and Zn2+ in low pH environments.[41] The potential establishment of white-rot fungi as a stable mycoremediator remains an important future discovery. White-rot fungi remain an important source of great unrealized potential.
Induced fungal decay
[edit]
A special way of giving grown wood an unusual structure is to infect it with a parasitic fungus by storing it in a humid environment (fungal decay). The fungus penetrates the layers of the wood and changes the nature of the cells. This process creates individual patterns and shades of colour. The wood treated in this way is then excellently suited for the production of all kinds of design objects. In order to stabilise the wood structure weakened by the fungus, resins or plastics are usually introduced into the material by special vacuum processes. This also kills the residual fungus after the desired pattern has been achieved, thus preserving the wood from being further consumed by the fungus.[42]
A special icing process applied to beech wood leads to results similar to those obtained with fungal decay. After the wood has been soaked, it is iced and then dried. The result is a very light wood with an almost black grain. This result, which also occurs very rarely in nature, is called ice-beech.[43][44]
Natural durability
[edit]Natural durability is the inherent capability of wood to tolerate and resist fungal decay and insect attacks, such as woodboring beetles and termites, and marine organisms.[45] This protective feature is attributable to specific biological compounds, called extractives that are toxic to wood-destroying organisms. Along with the tree’s growth, the sapwood converts into heartwood and this brings physical and chemical changes to the wood.[46] As a result, the permeability decreases while the natural durability increases. Thus, the extractives responsible for natural durability are mainly present in the heartwood, although they may also be contained in small amounts in the sapwood.[47] Different chemicals have been isolated from the heartwood of naturally rot-resistant trees and have shown to be protectants, including polyphenols, lignans (e.g. gmelinol, plicatic acid), flavonoids (e.g. mesquitol), tropolones (e.g. hinokitiol and other thujaplicins), sesquiterpenoids (e.g. α-cadinol).[48][49] The natural durability varies between tree species, geographic regions, environmental conditions, growth stage, and increases with the age. Thereby, some trees are more resistant to fungal diseases and insects and their timber lasts longer than other trees. Notably, the timber of these trees remain durable for a long-time period, even around a century, thereby they have been used as a reliable building material for centuries. Since the young trees do not produce enough protecting chemicals, some trees grow with a hollow, rotten trunk at an early age.[50] However, the stands of old-growth trees are more naturally durable than second-growth stands.[51] Tree species that have significant natural durability include Lagarostrobos franklinii (Huon pine), Intsia bijuga (ipil), some Eucalyptus species (ironbark), Podocarpus totara (totara), Vitex lucens (puriri), Agathis australis (kauri), and trees of the Cupressaceae family, such as Chamaecyparis obtusa (Hinoki cypress), Thuja plicata (Western red cedar), Thujopsis dolabrata (Hinoki asunaro), Juniperus cedrus (Canary Islands juniper), Cedrus atlantica (Atlas cedar), Chamaecyparis taiwanensis (Taiwan cypress), among others.[52]
According to the EN 350:2016 standards by the APA – The Engineered Wood Association, the durability of wood and wood-based products to fungal decay can be classified into five categories: very durable (DC1); durable (DC2); moderately durable (DC3); slightly durable (DC4); and not durable (DC5). The durability to insect attacks can be categorized as durable (DC D); moderately durable (DC M); and not durable (DC S).[45] Generally, the heartwood of durable tree species is considered as very durable, whereas the sapwood of all tree species is considered as not durable and is the most vulnerable.[citation needed]
Wood preservation
[edit]A wide selection of timber preservation has been developed to give the wood an improved durability and to protect it from decay. The wood can be treated according to the purpose (biological protection, e.g. fungi, insects, marine organisms) and the environment (interior, exterior, above ground, in ground, in water) of its use.[53] Timber preservatives include chromated copper arsenate (CCA), alkaline copper quaternary (ACQ), copper azole (CuAz), borates, sodium and potassium silicate, oil-based preservatives, such as creosote and pentachlorophenol, light organic solvent preservatives (LOSP), propiconazole-tebuconazole-imidacloprid, epoxy resins, wood acetylation, natural or biological preservation, such as treatment with heat (thermally modified wood), mud, tung oil, impregnation using biopolymers from agricultural waste (biological modified timber), covering wood with copper sheathes, etc. Treatment of timber with natural extractives derived from rot-resistant trees, such as hinokitiol, tannins, and tree extracts, is another promising environmentally-friendly wood preservation method.[54][55][56][57][58] The more permeable is the wood, the easier is it to treat. According to the EN 350:2016 standards, the treatability of woods can be categorized in four levels: (1) easy to treat; (2) moderately easy to treat; (3) difficult to treat; and (4) extremely difficult to treat.[45]
Safety
[edit]Over the years a lot of concerns have arisen regarding the arsenic and chromium contents of CCA. In 1986, the U.S. Environmental Protection Agency (EPA) recognized arsenic as a human carcinogen.[59] Water contamination with arsenic and its compounds is a serious public health issue, and their release to the environment and soil pollution is another environmental problem.[60][61] Different regulatory interventions have been undertaken worldwide to restrict their use in the wood industry, especially in timber for residential use. By the end of 2003, the U.S EPA and the wood industry agreed to discontinue the use of CCA in treating timber for residential use.[62] Its use is also prohibited in Canada, Australia, and the European Union.[63][64][65]
See also
[edit]
References
[edit]- ^ Harris, Samuel Y. (2001). Building Pathology: Deterioration, Diagnostics, and Intervention. John Wiley & Sons. p. 106. ISBN 978-0-471-33172-8.
- ^ "Wood Decay in Living and Dead Trees: A Pictorial Overview" (PDF). Archived from the original (PDF) on 2022-01-24. Retrieved 2018-02-28.
- ^ Viitanen, T. et al. (2010). Towards modelling of decay risk of wooden materials. European Journal of Wood and Wood Products 68:303-313.
- ^ a b c d J. Deacon, Wood decay and wood-rotting fungi. University of Edinburgh (2005?).
- ^ a b Microorganisms causing decay in trees and wood Archived 2020-02-20 at the Wayback Machine. University of Minnesota.
- ^ a b c Vane, C. H., et al. (2005). "Decay of cultivated apricot wood (Prunus armeniaca) by the ascomycete Hypocrea sulphurea, using solid state 13C NMR and off-line TMAH thermochemolysis with GC–MS." International Biodeterioration & Biodegradation 55(3): 175-185.
- ^ Olsson, Jörgen (2008). Colonization Patterns of Wood-inhabiting Fungi in Boreal Forest (PhD thesis). Umeå University.
- ^ Shingo Miyauchi, Hayat Hage, Elodie Drula, Laurence Lesage-Meessen, Jean-Guy Berrin, David Navarro, Anne Favel, Delphine Chaduli, Sacha Grisel, Mireille Haon, François Piumi, Anthony Levasseur, Anne Lomascolo, Steven Ahrendt, Kerrie Barry, Kurt M LaButti, Didier Chevret, Chris Daum, Jérôme Mariette, Christophe Klopp, Daniel Cullen, Ronald P de Vries, Allen C Gathman, Matthieu Hainaut, Bernard Henrissat, Kristiina S Hildén, Ursula Kües, Walt Lilly, Anna Lipzen, Miia R Mäkelä, Angel T Martinez, Mélanie Morel-Rouhier, Emmanuelle Morin, Jasmyn Pangilinan, Arthur F J Ram, Han A B Wösten, Francisco J Ruiz-Dueñas, Robert Riley, Eric Record, Igor V Grigoriev, Marie-Noëlle Rosso (2020). "Conserved white-rot enzymatic mechanism for wood decay in the Basidiomycota genus Pycnoporus". DNA Research. 27 (2). doi:10.1093/dnares/dsaa011. PMC 7406137. PMID 32531032.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jenna, Purhonen; Nerea, Abrego; Atte, Komonen; Seppo, Huhtinen; Heikki, Kotiranta; Thomas, Læssøe; Panu, Halme (2021-07-16). "Wood-inhabiting fungal responses to forest naturalness vary among morpho-groups". Scientific Reports. 11 (1): 14585. Bibcode:2021NatSR..1114585J. doi:10.1038/s41598-021-93900-7. hdl:10138/332607. ISSN 2045-2322. PMC 8285386. PMID 34272417.
- ^ Schilling, Marion; Farine, Sibylle; Péros, Jean-Pierre; Bertsch, Christophe; Gelhaye, Eric (2021-01-01), Morel-Rouhier, Mélanie; Sormani, Rodnay (eds.), "Chapter Six - Wood degradation in grapevine diseases", Advances in Botanical Research, Wood Degradation and Ligninolytic Fungi, vol. 99, Academic Press, pp. 175–207, doi:10.1016/bs.abr.2021.05.007, S2CID 238920143, retrieved 2023-03-29
- ^ Stamets, Paul (2005). Mycelium running: how mushrooms can help save the world. Random House, Inc. pp. 83–84. ISBN 978-1-58008-579-3.
- ^ a b Vane, C. H., et al. (2001). "The effect of fungal decay (Agaricus bisporus) on wheat straw lignin using pyrolysis–GC–MS in the presence of tetramethylammonium hydroxide (TMAH)." Journal of Analytical and Applied Pyrolysis 60(1): 69-78.
- ^ Ryvarden, Leif (1993). "Tropical polypores". In Isaac, Susan (ed.). Aspects of Tropical Mycology. British Mycological Society. Symposium. Cambridge University Press. p. 159. ISBN 978-0-521-45050-8.
- ^ a b Vane, C. H., et al. (2006). "Bark decay by the white-rot fungus Lentinula edodes: Polysaccharide loss, lignin resistance and the unmasking of suberin." International Biodeterioration & Biodegradation 57(1): 14-23.
- ^ Hoff JA, Klopfenstein NB, McDonald GI, Tonn JR, Kim MS, Zambino PJ, et al. (2004). "Fungal Endophytes in Woody Roots of Douglas-Fir (Pseudotsuga menziesii) and Ponderosa Pine (Pinus ponderosa)". Forest Pathology. 34 (4): 255–271. CiteSeerX 10.1.1.180.5697. doi:10.1111/j.1439-0329.2004.00367.x. ISSN 1437-4781. Retrieved 22 December 2022 – via U.S. Department of Agriculture Forest Service.
- ^ a b c Martinez, D., Larrondo, L. F., Putnam, N., Gelpke, M. D. S., Huang, K., Chapman, J., Helfenbein, K. G., Ramaiya, P., Detter, J. C., Larimer, F., Coutinho, P. M., Henrissat, B., Berka, R., Cullen, D., & Rokhsar, D. (2004). Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nature Biotechnology, 22(6), 695–700.
- ^ Palmer, J. M., & Evans, C. S. (1983). The Enzymic Degradation of Lignin by White-Rot Fungi. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 300(1100), 293–303.
- ^ Cohen, R.; Persky, L.; Hadar, Y. (2002). "Biotechnological applications and potential of wood-degrading mushrooms of the genus Pleurotus". Applied Microbiology and Biotechnology. 58 (5): 582–94. doi:10.1007/s00253-002-0930-y. PMID 11956739. S2CID 45444911.
- ^ a b Wymelenberg AV, Sabat G, Mozuch M, Kersten PJ, Cullen D, Blanchette RA (2006). "Structure, Organization, and Transcriptional Regulation of a Family of Copper Radical Oxidase Genes in the Lignin-Degrading Basidiomycete Phanerochaete chrysosporium". Applied and Environmental Microbiology. 72 (7): 4871–4877. Bibcode:2006ApEnM..72.4871V. doi:10.1128/AEM.00375-06. PMC 1489383. PMID 16820482.
- ^ Bissaro, B., Røhr, Å. K., Müller, G., Chylenski, P., Skaugen, M., Forsberg, Z., & Eijsink, V. G. (2017). Oxidative Cleavage of Polysaccharides by Copper Enzymes Depends on H2O2. Nature Chemical Biology, 13(10), 1123-1128.
- ^ Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, et al. (2009). Dixon RA (ed.). "Genome, Transcriptome, and Secretome Analysis of Wood Decay Fungus Postia placenta Supports Unique Mechanisms of Lignocellulose Conversion". Proceedings of the National Academy of Sciences of the United States of America. 106 (6): 1954–1959. Bibcode:2009PNAS..106.1954M. doi:10.1073/pnas.0809575106. PMC 2644145. PMID 19193860.
- ^ Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, et al. (2012). "The Paleozoic Origin of Enzymatic Lignin Decomposition Reconstructed from 31 Fungal Genomes". Science. 336 (6089): 1715–1719. Bibcode:2012Sci...336.1715F. doi:10.1126/science.1221748. hdl:10261/60626. PMID 22745431. S2CID 37121590.
- ^ Bogan, B. W., Schoenike, B., Lamar, R. T., & Cullen, D. (1996). Manganese Peroxidase mRNA and Enzyme Activity Levels During Bioremediation of Polycyclic Aromatic Hydrocarbon-Contaminated Soil with Phanerochaete chrysosporium. Applied and Environmental Microbiology, 62(7), 2381-2386.
- ^ Pogni R, Baratto MC, Teutloff C, Giansanti S, Ruiz-Dueñas FJ, Choinowski T, et al. (2006). "A Tryptophan Neutral Radical in the Oxidized State of Versatile Peroxidase from Pleurotus eryngii: A Combined Multifrequency EPR and Density Functional Theory Study". Journal of Biological Chemistry. 281 (14): 9517–9526. doi:10.1074/jbc.M510424200. PMID 16443605.
- ^ Riley, R., Salamov, A. A., Brown, D. W., Nagy, L. G., Floudas, D., Held, B. W., & Grigoriev, I. V. (2014). Extensive Sampling of Basidiomycete Genomes Demonstrates Inadequacy of the White-rot/brown-rot Paradigm for Wood Decay Fungi. Proceedings of the National Academy of Sciences, 111(27), 9923-9928.
- ^ a b Frandsen KE, Simmons TJ, Dupree P, Poulsen JN, Hemsworth GR, Ciano L, et al. (2016). "The Molecular Basis of Polysaccharide Cleavage by Lytic Polysaccharide Monooxygenases". Nature Chemical Biology. 12 (4): 298–303. doi:10.1038/nchembio.2029. PMC 4817220. PMID 26928935.
- ^ Brady, S. K., Sreelatha, S., Feng, Y., Chundawat, S. P., & Lang, M. J. Cellobiohydrolase 1 from Trichoderma reesei Degrades Cellulose in Single Cellobiose Steps. Nature Communications 6, 10149 (2015).
- ^ Ademark, P., Varga, A., Medve, J., Harjunpää, V., Drakenberg, T., Tjerneld, F., & Stålbrand, H. (1998). Softwood Hemicellulose-Degrading Enzymes from Aspergillus niger: Purification and Properties of a β-mannanase. Journal of Biotechnology, 63(3), 199-210.
- ^ Agger JW, Isaksen T, Várnai A, Vidal-Melgosa S, Willats WG, Ludwig R, et al. (2014). "Discovery of LPMO Activity on Hemicelluloses Shows the Importance of Oxidative Processes in Plant Cell Wall Degradation". Proceedings of the National Academy of Sciences. 111 (17): 6287–6292. Bibcode:2014PNAS..111.6287A. doi:10.1073/pnas.1323629111. PMC 4035949. PMID 24733907.
- ^ a b Lang, E., Eller, G., & Zadrazil, F. (1997). Lignocellulose Decomposition and Production of Ligninolytic Enzymes during Interaction of White Rot Fungi with Soil Microorganisms. Microbial Ecology, 34(1), 1–10.
- ^ a b Owens EM, Reddy CA, Grethlein HE (1994). "Outcome of interspecific interactions among brown-rot and white-rot wood decay fungi". FEMS Microbiology Ecology. 14 (1): 19–24. Bibcode:1994FEMME..14...19O. doi:10.1111/j.1574-6941.1994.tb00086.x.
- ^ a b Fukasawa Y, Gilmartin EC, Savoury M, Boddy L (2020). "Inoculum volume effects on competitive outcome and wood decay rate of brown-and white-rot basidiomycetes" (PDF). Fungal Ecology. 45: 100938. doi:10.1016/j.funeco.2020.100938. S2CID 216224049. 100938.
- ^ McClaugherty, C. A., Pastor, J., Aber, J. D., & Melillo, J. M. (1985). Forest Litter Decomposition in Relation to Soil Nitrogen Dynamics and Litter Quality. Ecology, 66(1), 266–275.
- ^ Folman LB, Klein Gunnewiek PJ, Boddy L, De Boer W (2008). "Impact of white-rot fungi on numbers and community composition of bacteria colonizing beech wood from forest soil". FEMS Microbiology Ecology. 63 (2): 181–191. Bibcode:2008FEMME..63..181F. doi:10.1111/j.1574-6941.2007.00425.x. PMID 18199083.
- ^ Floudas, D., Binder, M., Riley, R., Barry, K., Blanchette, R. A., Henrissat, B., Martínez, A.T., Otillar, R., Spatafora, J. W., Yadav, J. Y., Aerts, A., Benoit, I., Boyd, A., Carlson, A., Copeland, A., Coutinho, P. M., de Vries, R. P., Ferreira, P., Findley, K., & Hibbett, D. S. (2012). The Paleozoic Origin of Enzymatic Lignin Decomposition Reconstructed from 31 Fungal Genomes. Science, 336(6089), 1715–1719.
- ^ Kohler, A., Kuo, A., Nagy, L. G., Morin, E., Barry, K. W., Buscot, F., ... & Martin, F. (2015). Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nature genetics, 47(4), 410-415.
- ^ Ayuso-Fernández, I., Martínez, A. T., & Ruiz-Dueñas, F. J. (2017). Experimental recreation of the evolution of lignin-degrading enzymes from the Jurassic to date. Biotechnology for biofuels, 10(1), 1-13.
- ^ Ayuso-Fernández, I., Ruiz-Dueñas, F. J., & Martínez, A. T. (2018). Evolutionary convergence in lignin-degrading enzymes. Proceedings of the National Academy of Sciences, 115(25), 6428-6433.
- ^ Nagy, L. G., Riley, R., Bergmann, P. J., Krizsán, K., Martin, F. M., Grigoriev, I. V., ... & Hibbett, D. S. (2017). Genetic bases of fungal white rot wood decay predicted by phylogenomic analysis of correlated gene-phenotype evolution. Molecular biology and evolution, 34(1), 35-44.
- ^ Vane, C. H. (2003). "Monitoring Decay of Black Gum Wood (Nyssa sylvatica) During Growth of the Shiitake Mushroom (Lentinula edodes) Using Diffuse Reflectance Infrared Spectroscopy." Applied Spectroscopy 57(5): 514–517.
- ^ Arıca, M. Y., Bayramoǧlu, G., Yılmaz, M., Bektaş, S., & Genç, Ö. (2004). Biosorption of Hg2+, Cd2+, and Zn2+ by Ca-Alginate and Immobilized Wood-Rotting fungus Funalia trogii. Journal of Hazardous Materials, 109(1-3), 191-199.
- ^ "Gestocktes Holz". mortalitas.eu. February 2016. Retrieved 2020-03-31.
- ^ "Eisbuche". eisbuche.de. Retrieved 2020-03-31.
- ^ "Mit Hilfe von Väterchen Frost". bm-online.de. 6 November 2018. Retrieved 2020-03-31.
- ^ a b c "EN 350: 2016 (updated) | APAwood - Europe". APA – The Engineered Wood Association. 17 August 2022.
- ^ Verbist, Maxime; Nunes, Lina; Jones, Dennis; Branco, Jorge M. (2019). "Service life design of timber structures". Long-term Performance and Durability of Masonry Structures: 311–336. doi:10.1016/B978-0-08-102110-1.00011-X. ISBN 9780081021101. S2CID 116669346.
- ^ Munir, Muhammad Tanveer; Pailhories, Hélène; Eveillard, Matthieu; Irle, Mark; Aviat, Florence; Federighi, Michel; Belloncle, Christophe (24 August 2020). "Experimental Parameters Influence the Observed Antimicrobial Response of Oak Wood (Quercus petraea)". Antibiotics. 9 (9): 535. doi:10.3390/antibiotics9090535. PMC 7558063. PMID 32847132.
- ^ Singh, Tripti; Singh, Adya P. (September 2012). "A review on natural products as wood protectant". Wood Science and Technology. 46 (5): 851–870. doi:10.1007/s00226-011-0448-5. S2CID 16934998.
- ^ Morris, Paul I.; Stirling, Rod (September 2012). "Western red cedar extractives associated with durability in ground contact". Wood Science and Technology. 46 (5): 991–1002. doi:10.1007/s00226-011-0459-2. S2CID 15869687.
- ^ Cedar: tree of life to the Northwest Coast Indians. Vancouver, B.C.: Douglas & McIntyre. 1984. p. 22. ISBN 0-88894-437-3.
- ^ Scheffer, T. C.; Morrell, Jeffrey J.; Laboratory, Oregon State University Forest Research. "Natural durability of wood: a worldwide checklist of species". ir.library.oregonstate.edu. hdl:1957/7736.
- ^ Munir, Muhammad Tanveer; Pailhories, Hélène; Eveillard, Matthieu; Irle, Mark; Aviat, Florence; Dubreil, Laurence; Federighi, Michel; Belloncle, Christophe (1 May 2020). "Testing the Antimicrobial Characteristics of Wood Materials: A Review of Methods". Antibiotics. 9 (5): 225. doi:10.3390/antibiotics9050225. PMC 7277147. PMID 32370037.
- ^ Woodard, A.C.; Milner, H.R. (2016). "Sustainability of timber and wood in construction". Sustainability of Construction Materials: 129–157. doi:10.1016/B978-0-08-100370-1.00007-X. ISBN 9780081009956.
- ^ Silveira, Amanda G. Da; Santini, Elio J.; Kulczynski, Stela M.; Trevisan, Rômulo; Wastowski, Arci D.; Gatto, Darci A. (7 December 2017). "Tannic extract potential as natural wood preservative of Acacia mearnsii". Anais da Academia Brasileira de Ciências. 89 (4): 3031–3038. doi:10.1590/0001-3765201720170485. PMID 29236851.
- ^ Syofuna, A; Banana, A.Y; Nakabonge, G (2012). "Efficiency of natural wood extractives as wood preservatives against termite attack". Maderas. Ciencia y tecnología. 14 (2): 155–163. doi:10.4067/S0718-221X2012000200003.
- ^ Binbuga, Nursen; Ruhs, Christopher; Hasty, Julia K.; Henry, William P.; Schultz, Tor P. (1 May 2008). "Developing environmentally benign and effective organic wood preservatives by understanding the biocidal and non-biocidal properties of extractives in naturally durable heartwood". Holzforschung. 62 (3): 264–269. doi:10.1515/HF.2008.038. S2CID 97166844.
- ^ Hu, Junyi; Shen, Yu; Pang, Song; Gao, Yun; Xiao, Guoyong; Li, Shujun; Xu, Yingqian (December 2013). "Application of hinokitiol potassium salt for wood preservative". Journal of Environmental Sciences. 25: S32–S35. doi:10.1016/S1001-0742(14)60621-5. PMID 25078835.
- ^ Brocco, Victor Fassina; Paes, Juarez Benigno; Costa, Lais Gonçalves da; Brazolin, Sérgio; Arantes, Marina Donária Chaves (January 2017). "Potential of teak heartwood extracts as a natural wood preservative". Journal of Cleaner Production. 142: 2093–2099. doi:10.1016/j.jclepro.2016.11.074.
- ^ US EPA, ORD. "Arsenic, Inorganic CASRN 7440-38-2 | DTXSID4023886 | IRIS | US EPA, ORD". cfpub.epa.gov.
- ^ "Arsenic". www.who.int. World Health Organization.
- ^ Belluck, D. A.; Benjamin, S. L.; Baveye, P.; Sampson, J.; Johnson, B. (March 2003). "Widespread Arsenic Contamination of Soils in Residential Areas and Public Spaces: An Emerging Regulatory or Medical Crisis?". International Journal of Toxicology. 22 (2): 109–128. doi:10.1080/10915810305087. PMID 12745992. S2CID 20986621.
- ^ "Response to Requests to Cancel Certain Chromated Copper Arsenate (CCA) Wood Preservative Products and Amendments to Terminate Certain Uses of other CCA Products". Federal Register. 9 April 2003.
- ^ Canada, Environment and Climate Change (26 February 2014). "Wood preservation facilities, chromated copper arsenate: chapter B-1". Government of Canada.
- ^ "New restrictions in place for arsenic-treated timber". Australian Pesticides and Veterinary Medicines Authority. 22 September 2014.
- ^ "EUR-Lex - 32003L0002 - EN - EUR-Lex". Eur-lex.europa.eu.
Further reading
[edit]- Schwarze, Francis W. M. R.; Engels, Julia; Mattheck, Claus (2000). Fungal Strategies of Wood Decay in Trees. Springer. ISBN 978-3-540-67205-0.
- Mycorrhizal fungi and soil carbon storage
- White, Robert H.; Ross, Robert J. (November 2014). Wood and Timber Condition Assessment Manual (2nd ed.). Madison, WI: United States Department of Agriculture, Forest Service, Forest Products Laboratory. Retrieved 31 January 2015.
- Wasser, Zmitrovich I. V.; Engels, Tura (2014). Wood-inhabiting fungi (PDF). Fungi from different substrates / J. K. Misra, J. P. Tewari, S. K. Deshmukh, C. Vágvölgyi (eds). N. Y.: CRC Press, Taylor and Francis group.


