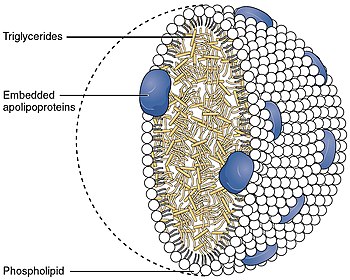
Apolipoprotein B
Apolipoprotein B (ApoB) is a protein that in humans is encoded by the APOB gene. Its measurement is commonly used to detect risk of atherosclerotic cardiovascular disease.[5][6]
Function
[edit]Apolipoprotein B is the primary apolipoprotein of chylomicrons, VLDL, Lp(a), IDL, and LDL particles (LDL—commonly known as "bad cholesterol" when in reference to both heart disease and vascular disease in general), which is responsible for carrying fat molecules (lipids), including cholesterol, around the body to all cells within all tissues. While all the functional roles of ApoB within the LDL (and all larger) particles remain somewhat unclear, it is the primary organizing protein (of the entire complex shell enclosing/carrying fat molecules within) component of the particles and is absolutely required for the formation of these particles. What is also clear is that the ApoB on the LDL particle acts as a ligand for LDL receptors in various cells throughout the body (i.e., less formally, ApoB indicates fat-carrying particles are ready to enter any cells with ApoB receptors and deliver fats carried within into the cells).
Through mechanisms only partially understood, high levels of ApoB, especially associated with the higher LDL particle concentrations, are the primary driver of plaques that cause vascular disease (atherosclerosis), commonly first becoming obviously symptomatic as heart disease, stroke and many other body wide complications after decades of progression. There is considerable evidence that concentrations of ApoB[7][8] and especially the NMR assay[9] (specific for LDL-particle concentrations) are superior indicators of vascular/heart disease driving physiology than either total cholesterol or LDL-cholesterol (as long promoted by the NIH starting in the early 1970s). However, primarily for historic cost/complexity reasons, cholesterol, and estimated LDL-cholesterol by calculation, remains the most commonly promoted lipid test for the risk factor of atherosclerosis. ApoB is routinely measured using immunoassays such as ELISA or nephelometry. Refined and automated NMR methods allow measurement distinctions between the many different ApoB particles.
Genetic disorders
[edit]High levels of ApoB are related to heart disease. Hypobetalipoproteinemia is a genetic disorder that can be caused by a mutation in the ApoB gene, APOB.[10] Abetalipoproteinaemia is usually[vague] caused by a mutation in the MTP gene, MTP.[11]
Mutations in gene APOB100 can also cause familial hypercholesterolemia,[12] a hereditary (autosomal dominant) form of metabolic disorder hypercholesterolemia.
Mouse studies
[edit]Mice have been used as model organisms in ApoB study as they express an equivalent protein known as mouse ApoB (mApoB). Mice overexpressing mApoB have increased levels of LDL and decreased levels of HDL.[13] Mice containing only one functional copy of the mApoB gene show the opposite effect, being resistant to hypercholesterolemia. Mice containing no functional copies of the gene are not viable.[14]
Molecular biology
[edit]The protein occurs in the plasma in two main isoforms, ApoB48 and ApoB100. The first is synthesized exclusively by the small intestine, the second by the liver.[15] ApoB-100 is the largest of the apoB group of proteins, consisting of 4563 amino acids, including a 27-amino acid signal peptide and a 4536-amino acid mature protein.[15] Both isoforms are coded by APOB and by a single mRNA transcript larger than 16 kb. ApoB48 is generated when a stop codon (UAA) at residue 2153 is created by RNA editing. There appears to be a trans-acting tissue-specific splicing gene that determines which isoform is ultimately produced.[citation needed] Alternatively, there is some evidence that a cis-acting element several thousand bp upstream determines which isoform is produced.[citation needed]
As a result of the RNA editing, ApoB48 and ApoB100 share a common N-terminal sequence, but ApoB48 lacks ApoB100's C-terminal LDL receptor binding region. In fact, ApoB48 is so-called because it constitutes 48% of the sequence for ApoB100.
ApoB 48 is a unique protein to chylomicrons from the small intestine. After most of the lipids in the chylomicron have been absorbed, ApoB48 returns to the liver as part of the chylomicron remnant, where it is endocytosed and degraded.
Clinical significance
[edit]Benefits
[edit]Role in innate immune system
[edit]Very low-density lipoproteins and low-density lipoproteins interfere with the quorum sensing system that upregulates genes required for invasive Staphylococcus aureus infection. The mechanism of antagonism entails binding ApoB, to a S. aureus autoinducer pheromone, preventing signaling through its receptor. Mice deficient in ApoB are more susceptible to invasive bacterial infection.[16]
Adverse effects
[edit]Role in insulin resistance
[edit]Overproduction of apolipoprotein B can result in lipid-induced endoplasmic reticulum stress and insulin resistance in the liver.[17]
Role in lipoproteins and atherosclerosis
[edit]ApoB100 is found in lipoproteins originating from the liver (VLDL, IDL, LDL[18]). Importantly, there is one ApoB100 molecule per hepatic-derived lipoprotein. Hence, using that fact, one can quantify the number of lipoprotein particles by noting the total ApoB100 concentration in the circulation. Since there is one and only one ApoB100 per particle, the number of particles is reflected by the ApoB100 concentration. The same technique can be applied to individual lipoprotein classes (e.g. LDL) and thereby enable one to count them as well.
It is well established that ApoB100 levels are associated with coronary heart disease, they are a far better predictor of it than are LDL-C concentrations.[19][20][21] Reason: LDL-C does not reflect actual particle concentrations & cholesterol cannot dissolve or move (in water) without particles to carry it. A simple way to understand this observation is the fact that ApoB100, one per particle, reflects actual lipoprotein particle concentration (independent of their cholesterol, or other lipid content). In this way, one can understand that the number of ApoB100-containing lipoprotein particles which can carry lipids into the artery walls is a key determinant, driver of atherosclerosis and heart disease.
One way to explain the above is to consider that large numbers of lipoprotein particles, and, in particular, large numbers of LDL particles, lead to competition at the ApoB100 receptor (i.e. LDL receptor) of peripheral cells. Since such competition will prolong the residence time of LDL particles in the circulation, it may lead to greater opportunity for them to undergo oxidation and/or other chemical modifications. Such modifications may lessen the particles' ability to be cleared by the classic LDL receptor and/or increase their ability to interact with so-called "scavenger" receptors. The net result is the shunting of LDL particles to these scavenger receptors. Scavenger receptors typically are found on macrophages, with cholesterol-laden macrophages being better known as "foam cells". Foam cells characterize atherosclerotic lesions. In addition to this possible mechanism of foam cell generation, an increase in the levels of chemically modified LDL particles may also lead to an increase in endothelial damage. This occurs as a result of modified-LDL's toxic effect on vascular endothelium as well as its ability both to recruit immune effector cells and to promote platelet activation.
The INTERHEART study found that the ApoB100 / ApoA1 ratio is more effective at predicting heart attack risk, in patients who had had an acute myocardial infarction, than either the ApoB100 or ApoA1 measure alone.[22] (ApoA1 is the major HDL protein.[23]) In the general population this remains unclear although in a recent study ApoB was the strongest risk marker for cardiovascular events.[24]
A Mediterranean diet is recommended as a means of lowering Apolipoprotein B.[25]
Interactions
[edit]ApoB has been shown to interact with apo(a),[26] PPIB,[27] Calcitonin receptor[27][28] and HSP90B1.[27][28] Interaction of ApoB with proteoglycans, collagen, and fibronectin is believed to cause atherosclerosis.[29][30]
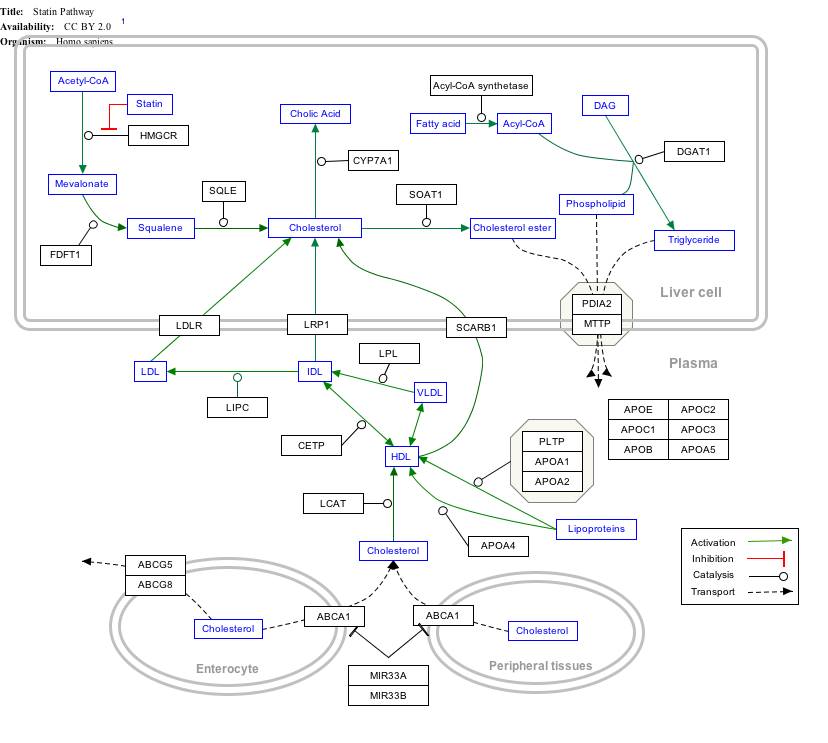
Interactive pathway map
[edit]Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- ^ The interactive pathway map can be edited at WikiPathways: "Statin_Pathway_WP430".
Regulation
[edit]The expression of APOB is regulated by cis-regulatory elements in the APOB 5′ UTR and 3′ UTR.[31]
RNA editing
[edit]The mRNA encoding this protein is subject to cytidine to uridine (C to U) site-specific RNA editing. ApoB100 and ApoB48 are encoded by the same gene, however, the differences in the translated proteins are not due to alternative splicing but are due to the tissue-specific RNA editing event. ApoB mRNA editing was the first example of editing observed in vertebrates.[32] Editing of ApoB mRNA occurs in all placental mammals.[33] Editing occurs post transcriptionally as the nascent polynucleotides do not contain edited nucleosides.[34]
Type
[edit]C to U editing of ApoB mRNA requires an editing complex or holoenzyme (editosome) consisting of the C to U-editing enzyme Apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (ApoBEC-1) as well as other auxiliary factors. ApoBEC-1 is a protein that in humans is encoded by the APOBEC1 gene.[35][1]It is a member of the cytidine deaminase family. ApoBEC-1 alone is not sufficient for the editing of ApoB mRNA [36] and requires at least one of these auxiliary factors, APOBEC1 complementation factor (A1CF)[37] for editing to occur. A1CF contains 3 non identical repeats. It acts as the RNA binding subunit and directs ApoBEC-1 to the ApoB mRNA downstream of the edited cytidine.[38] Other auxiliary factors are known to be part of the holoenzyme. Some of these proteins have been identified. these are CUG binding protein 2 (CUGBP2),[39] SYNCRIP (glycine-arginine-tyrosine-rich RNA binding protein, GRY-RBP),[40] heterogeneous nuclear ribonucleoprotein (hnRNP)-C1 (HNRNPC),[41] ApoBEC-1 binding protein ABBP1 (HNRNPAB), ABBP2,[42] KH-type splicing regulatory binding protein (KHSRP), Bcl-2-associated athanogene 4 (BAG4),[43] and auxiliary factor (AUX)240.[44] All these proteins have been identified using detection assays and have all been demonstrated to interact with either ApoBEC-1, A1CF, or ApoB RNA. The function of these auxiliary proteins in the editing complex are unknown. As well as editing ApoB mRNA, the ApoBEC-1 editsome also edits the mRNA of NF1. mRNA editing of ApoB mRNA is the best defined example of this type of C to U RNA editing in humans.
Location
[edit]Despite being a 14,000 residue long transcript, a single cytidine is targeted for editing. Within the ApoB mRNA a sequence consisting of 26 nucleotides necessary for editing is found. This is known as the editing motif. These nucleotides (6662–6687) were determined to be essential by site specific mutagenesis experiments.[45] An 11 nucleotide portion of this sequence 4–5 nucleotides downstream from the editing site is an important region known as the mooring sequence.[46] A region called the spacer element is found 2–8 nucleotides between the edited nucleoside and this mooring sequence.[47] There is also a regulatory sequence 3′ to the editing site. The active site of ApoBEC-1, the catalytic component of the editing holoenzyme is thought to bind to an AU rich region of the mooring sequence with the aid of ACF in binding the complex to the mRNA.[48] The edited cytidine residue is located at nucleotide 6666 located in exon 26 of the gene. Editing at this site results in a codon change from a Glutamine codon (CAA) to an inframe stop codon (UAA).[32] Computer modelling has detected for editing to occur, the edited Cytidine is located in a loop.[46] The selection of the edited cytidine is also highly dependent on this secondary structure of the surrounding RNA. There are also some indications that this loop region is formed between the mooring sequence and the 3′ regulatory region of the ApoB mRNA.[49] The predicted secondary structure formed by ApoB mRNA is thought to allow for contact between the residue to be edited and the active site of APOBEC1 as well as for binding of ACF and other auxiliary factors associated with the editosome.
Regulation
[edit]Editing of ApoB mRNA in humans is tissue regulated, with ApoB48 being the main ApoB protein of the small intestine in humans. It occurs in lesser amounts in the colon, kidney and stomach along with the non edited version.[50] Editing is also developmentally regulated with the non edited version only being translated early in development but the edited form increases during development in the tissues where editing can occur.[51][52] Editing levels of ApoB mRNA have been shown to vary in response to changes in diet. exposure to alcohol and hormone levels.[53][54][55]
Conservation
[edit]ApoB mRNA editing also occurs in mice, and rats. In contrast to humans editing occurs in liver in mice and rats up to a frequency of 65%.[56] It has not been observed in birds or lesser species.[57]
Consequences
[edit]Structure
[edit]Editing results in a codon change creating an in-frame stop codon leading to translation of a truncated protein, ApoB48. This stop codon results in the translation of a protein that lacks the carboxyl terminus which contains the protein's LDLR binding domain. The full protein ApoB100 which has nearly 4500 amino acids is present in VLDL and LDL. Since many parts of ApoB100 are in an amphipathic condition, the structure of some of its domains is dependent on underlying lipid conditions. However, it is known to have the same overall folding in LDL having five main domains. Recently the first structure of LDL at human body temperature in native condition has been found using cryo-electron microscopy at a resolution of 16 Angstrom.[58] The overall folding of ApoB-100 has been confirmed and some heterogeneity in the local structure of its domains have been mapped.[citation needed]
Function
[edit]Editing is restricted to those transcripts expressed in the small intestine. This shorter version of the protein has a function specific to the small intestine. The main function of the full length liver expressed ApoB100 is as a ligand for activation of the LDL-R. However, editing results in a protein lacking this LDL-R binding region of the protein. This alters the function of the protein and the shorter ApoB48 protein as specific functions relative to the small intestine. ApoB48 is identical to the amino-terminal 48% of ApoB100.[59] The function of this isoform is in fat absorption of the small intestine and is involved in the synthesis, assembly and secretion of chylomicrons. These chylomicrons transport dietary lipids to tissues while the remaining chylomicrons along with associated residual lipids are in 2–3 hours taken up by the liver via the interaction of apolipoprotein E (ApoE) with lipoprotein receptors. It is the dominant ApoB protein in the small intestine of most mammals. It is a key protein in the exogenous pathway of lipoprotein metabolism. Intestinal proteins containing ApoB48 are metabolized to chylomicron remnant particles which are taken up by remnant receptors.
See also
[edit]References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000084674 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000020609 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Glavinovic T, Thanassoulis G, de Graaf J, Couture P, Hegele RA, Sniderman AD (October 2022). "Physiological Bases for the Superiority of Apolipoprotein B Over Low-Density Lipoprotein Cholesterol and Non-High-Density Lipoprotein Cholesterol as a Marker of Cardiovascular Risk". Journal of the American Heart Association. 11 (20): e025858. doi:10.1161/JAHA.122.025858. PMC 9673669. PMID 36216435.
- ^ Behbodikhah J, Ahmed S, Elyasi A, Kasselman LJ, De Leon J, Glass AD, Reiss AB (October 2021). "Apolipoprotein B and Cardiovascular Disease: Biomarker and Potential Therapeutic Target". Metabolites. 11 (10): 690. doi:10.3390/metabo11100690. PMC 8540246. PMID 34677405.
- ^ Lim JS, Lee DH, Park JY, Jin SH, Jacobs DR (2011). "Reliability of low-density lipoprotein cholesterol, non-high-density lipoprotein cholesterol, and apolipoprotein B measurement". Journal of Clinical Lipidology. 5 (4): 264–272. doi:10.1016/j.jacl.2011.05.004. PMID 21784371.
- ^ Jacobson TA (2011). "Opening a new lipid "apo-thecary": incorporating apolipoproteins as potential risk factors and treatment targets to reduce cardiovascular risk". Mayo Clinic Proceedings. 86 (8): 762–780. doi:10.4065/mcp.2011.0128. PMC 3146376. PMID 21803958.
- ^ Carmena R, Duriez P, Fruchart JC (2004). "Atherosclerosis: Evolving Vascular Biology and Clinical Implications". Circulation. 109 (23): III–2. doi:10.1161/01.CIR.0000131511.50734.44. PMID 15198959.
- ^ Young, Stephen G.; Hubl, Susan T.; Chappell, David A.; Smith, Richard S.; Claiborne, Frederica; Snyder, Steven M.; Terdiman, Joseph F. (1989-06-15). "Familial Hypobetalipoproteinemia Associated with a Mutant Species of Apolipoprotein B (B-46)". New England Journal of Medicine. 320 (24): 1604–1610. doi:10.1056/NEJM198906153202407. ISSN 0028-4793. PMID 2725600.
- ^ "MTTP microsomal triglyceride transfer protein [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2024-03-28.
- ^ Andersen, Lars H.; Miserez, André R.; Ahmad, Zahid; Andersen, Rolf L. (November 2016). "Familial defective apolipoprotein B-100: A review". Journal of Clinical Lipidology. 10 (6): 1297–1302. doi:10.1016/j.jacl.2016.09.009. PMID 27919345.
- ^
 McCormick SP, Ng JK, Véniant M, Borén J, Pierotti V, Flynn LM, et al. (May 1996). "Transgenic mice that overexpress mouse apolipoprotein B. Evidence that the DNA sequences controlling intestinal expression of the apolipoprotein B gene are distant from the structural gene". The Journal of Biological Chemistry. 271 (20): 11963–11970. doi:10.1074/jbc.271.20.11963. PMID 8662599.
McCormick SP, Ng JK, Véniant M, Borén J, Pierotti V, Flynn LM, et al. (May 1996). "Transgenic mice that overexpress mouse apolipoprotein B. Evidence that the DNA sequences controlling intestinal expression of the apolipoprotein B gene are distant from the structural gene". The Journal of Biological Chemistry. 271 (20): 11963–11970. doi:10.1074/jbc.271.20.11963. PMID 8662599.
- ^
 Farese RV, Ruland SL, Flynn LM, Stokowski RP, Young SG (February 1995). "Knockout of the mouse apolipoprotein B gene results in embryonic lethality in homozygotes and protection against diet-induced hypercholesterolemia in heterozygotes". Proceedings of the National Academy of Sciences of the United States of America. 92 (5): 1774–1778. Bibcode:1995PNAS...92.1774F. doi:10.1073/pnas.92.5.1774. PMC 42602. PMID 7878058.
Farese RV, Ruland SL, Flynn LM, Stokowski RP, Young SG (February 1995). "Knockout of the mouse apolipoprotein B gene results in embryonic lethality in homozygotes and protection against diet-induced hypercholesterolemia in heterozygotes". Proceedings of the National Academy of Sciences of the United States of America. 92 (5): 1774–1778. Bibcode:1995PNAS...92.1774F. doi:10.1073/pnas.92.5.1774. PMC 42602. PMID 7878058.
- ^ a b Chen SH, Yang CY, Chen PF, Setzer D, Tanimura M, Li WH, Gotto AM Jr, Chan L (1986). "The complete cDNA and amino acid sequence of human apolipoprotein B-100". Journal of Biological Chemistry. 261 (28): 12918–12921. doi:10.1016/S0021-9258(18)69248-8. PMID 3759943.
- ^ Peterson MM, Mack JL, Hall PR, Alsup AA, Alexander SM, Sully EK, Sawires YS, Cheung AL, Otto M, Gresham HD (2008). "Apolipoprotein B is an innate barrier against invasive Staphylococcus aureus infection". Cell Host & Microbe. 4 (6): 507–509. doi:10.1016/j.chom.2008.10.001. PMC 2639768. PMID 19064256.
- ^ Su Q, Tsai J, Xu E, Qiu W, Bereczki E, Santha M, Adeli K (2009). "Apolipoprotein B100 acts as a molecular link between lipid-induced endoplasmic reticulum stress and hepatic insulin resistance". Hepatology. 50 (1): 77–84. doi:10.1002/hep.22960. PMID 19434737. S2CID 205869807.
- ^ MedlinePlus Encyclopedia: Apolipoprotein B100
- ^ Cromwell WC, Otvos JD, Keyes MJ, Pencina MJ, Sullivan L, Vasan RS, Wilson PW, D'Agostino RB (December 2007). "LDL Particle Number and Risk of Future Cardiovascular Disease in the Framingham Offspring Study - Implications for LDL Management". Journal of Clinical Lipidology. 1 (6): 583–592. doi:10.1016/j.jacl.2007.10.001. PMC 2720529. PMID 19657464.
- ^ Sniderman AD, Lamarche B, Contois JH, de Graaf J (December 2014). "Discordance analysis and the Gordian Knot of LDL and non-HDL cholesterol versus apoB". Current Opinion in Lipidology. 25 (6): 461–467. doi:10.1097/MOL.0000000000000127. PMID 25340478. S2CID 23464159.
- ^ Glavinovic T, Thanassoulis G, Sniderman AD (2022). "Physiological Bases for the Superiority of Apolipoprotein B Over Low-Density Lipoprotein Cholesterol and Non-High-Density Lipoprotein Cholesterol as a Marker of Cardiovascular Risk". Journal of the American Heart Association. 11 (20): e025858. doi:10.1161/JAHA.122.025858. PMC 9673669. PMID 36216435.
- ^ McQueen MJ, Hawken S, Wang X, Ounpuu S, Sniderman A, Probstfield J, Steyn K, Sanderson JE, Hasani M, Volkova E, Kazmi K, Yusuf S (July 2008). "Lipids, lipoproteins, and apolipoproteins as risk markers of myocardial infarction in 52 countries (the INTERHEART study): a case-control study". Lancet. 372 (9634): 224–233. doi:10.1016/S0140-6736(08)61076-4. PMID 18640459. S2CID 26567691.
- ^ van der Vorst EP (2020). "High-Density Lipoproteins and Apolipoprotein A1". Vertebrate and Invertebrate Respiratory Proteins, Lipoproteins and other Body Fluid Proteins. Subcellular Biochemistry. Vol. 94. pp. 399–420. doi:10.1007/978-3-030-41769-7_16. ISBN 978-3-030-41768-0. PMID 32189309. S2CID 213180689.
- ^ Benn M, Nordestgaard BG, Jensen GB, Tybjaerg-Hansen A (2007). "Improving prediction of ischemic cardiovascular disease in the general population using apolipoprotein B: the Copenhagen City Heart Study". Arterioscler Thromb Vasc Biol. 27 (3): 661–670. doi:10.1161/01.ATV.0000255580.73689.8e. PMID 17170368.
- ^ Lamantia V, Sniderman A, Faraj M (2016). "Nutritional management of hyperapoB". Nutrition Research Reviews. 29 (2): 202–233. doi:10.1017/S0954422416000147. PMID 27821191.
- ^ Malaguarnera M, Vacante M, Russo C, Malaguarnera G, Antic T, Malaguarnera L, Bella R, Pennisi G, Galvano F, Frigiola A (2013). "Lipoprotein(a) in cardiovascular diseases". BioMed Research International. 2013 (650989): 1–9. doi:10.1155/2013/650989. PMC 3591100. PMID 23484137.
- ^ a b c Zhang J, Herscovitz H (February 2003). "Nascent lipidated apolipoprotein B is transported to the Golgi as an incompletely folded intermediate as probed by its association with network of endoplasmic reticulum molecular chaperones, GRP94, ERp72, BiP, calreticulin, and cyclophilin B". J. Biol. Chem. 278 (9): 7459–7468. doi:10.1074/jbc.M207976200. PMID 12397072.
- ^ a b Linnik KM, Herscovitz H (Aug 1998). "Multiple molecular chaperones interact with apolipoprotein B during its maturation. The network of endoplasmic reticulum-resident chaperones (ERp72, GRP94, calreticulin, and BiP) interacts with apolipoprotein b regardless of its lipidation state". J. Biol. Chem. 273 (33): 21368–21373. doi:10.1074/jbc.273.33.21368. PMID 9694898.
- ^ Khalil MF, Wagner WD, Goldberg IJ (2004). "Lipoprotein(a) in cardiovascular diseases". Arteriosclerosis, Thrombosis, and Vascular Biology. 24 (12): 2211–2218. doi:10.1161/01.ATV.0000147163.54024.70. PMID 15472124.
- ^ Tabas I, Williams KJ, Borén J (2007). "Subendothelial lipoprotein retention as the initiating process in atherosclerosis: update and therapeutic implications". Circulation. 116 (16): 1832–1844. doi:10.1161/circulationaha.106.676890. PMID 17938300.
- ^ Pontrelli L, Sidiropoulos KG, Adeli K (2004). "Translational control of apolipoprotein B mRNA: regulation via cis elements in the 5′ and 3′ untranslated regions". Biochemistry. 43 (21): 6734–6744. doi:10.1021/bi049887s. PMID 15157107.
- ^ a b Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J (September 1987). "A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine". Cell. 50 (6): 831–840. doi:10.1016/0092-8674(87)90510-1. PMID 3621347. S2CID 37938313.
- ^ Fujino T, Navaratnam N, Jarmuz A, von Haeseler A, Scott J (July 1999). "C→U editing of apolipoprotein B mRNA in marsupials: identification and characterisation of APOBEC-1 from the American opossum Monodelphus domestica". Nucleic Acids Res. 27 (13): 2662–2671. doi:10.1093/nar/27.13.2662. PMC 148475. PMID 10373583.
- ^ Lau PP, Xiong WJ, Zhu HJ, Chen SH, Chan L (October 1991). "Apolipoprotein B mRNA editing is an intranuclear event that occurs posttranscriptionally coincident with splicing and polyadenylation". J. Biol. Chem. 266 (30): 20550–20554. doi:10.1016/S0021-9258(18)54960-7. PMID 1939106.
- ^ "APOBEC1 Gene - GeneCards | ABEC1 Protein | ABEC1 Antibody". Archived from the original on 2011-07-26. Retrieved 2011-02-24.
- ^ Navaratnam N, Fujino T, Bayliss J, Jarmuz A, How A, Richardson N, Somasekaram A, Bhattacharya S, Carter C, Scott J (January 1998). "Escherichia coli cytidine deaminase provides a molecular model for ApoB RNA editing and a mechanism for RNA substrate recognition". J. Mol. Biol. 275 (4): 695–714. doi:10.1006/jmbi.1997.1506. PMID 9466941.
- ^ "A1CF Gene - GeneCards | A1CF Protein | A1CF Antibody". Archived from the original on 2011-07-26. Retrieved 2011-02-24.
- ^ Blanc V, Kennedy S, Davidson NO (October 2003). "A novel nuclear localization signal in the auxiliary domain of apobec-1 complementation factor regulates nucleocytoplasmic import and shuttling". J. Biol. Chem. 278 (42): 41198–41204. doi:10.1074/jbc.M302951200. PMID 12896982.
- ^ Anant S, Henderson JO, Mukhopadhyay D, Navaratnam N, Kennedy S, Min J, Davidson NO (December 2001). "Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and A1CF, the apobec-1 complementation factor". J. Biol. Chem. 276 (50): 47338–47351. doi:10.1074/jbc.M104911200. PMID 11577082.
- ^ Blanc V, Navaratnam N, Henderson JO, Anant S, Kennedy S, Jarmuz A, Scott J, Davidson NO (March 2001). "Identification of GRY-RBP as an apolipoprotein B RNA-binding protein that interacts with both apobec-1 and apobec-1 complementation factor to modulate C to U editing". J. Biol. Chem. 276 (13): 10272–10283. doi:10.1074/jbc.M006435200. PMID 11134005.
- ^ Greeve J, Lellek H, Rautenberg P, Greten H (1998). "Inhibition of the apolipoprotein B mRNA editing enzyme-complex by hnRNP C1 protein and 40S hnRNP complexes". Biol. Chem. 379 (8–9): 1063–1073. doi:10.1515/bchm.1998.379.8-9.1063. PMID 9792439. S2CID 25911416.
- ^ Lau PP, Villanueva H, Kobayashi K, Nakamuta M, Chang BH, Chan L (December 2001). "A DnaJ protein, apobec-1-binding protein-2, modulates apolipoprotein B mRNA editing". J. Biol. Chem. 276 (49): 46445–46452. doi:10.1074/jbc.M109215200. PMID 11584023.
- ^ Lau PP, Chan L (December 2003). "Involvement of a chaperone regulator, Bcl2-associated athanogene-4, in apolipoprotein B mRNA editing". J. Biol. Chem. 278 (52): 52988–52996. doi:10.1074/jbc.M310153200. PMID 14559896.
- ^ Schock D, Kuo SR, Steinburg MF, Bolognino M, Sparks JD, Sparks CE, Smith HC (February 1996). "An auxiliary factor containing a 240-kDa protein complex is involved in apolipoprotein B RNA editing". Proc. Natl. Acad. Sci. U.S.A. 93 (3): 1097–1102. Bibcode:1996PNAS...93.1097S. doi:10.1073/pnas.93.3.1097. PMC 40037. PMID 8577721.
- ^ Davies MS, Wallis SC, Driscoll DM, Wynne JK, Williams GW, Powell LM, Scott J (August 1989). "Sequence requirements for apolipoprotein B RNA editing in transfected rat hepatoma cells". J. Biol. Chem. 264 (23): 13395–13398. doi:10.1016/S0021-9258(18)80008-4. PMID 2760026.
- ^ a b Shah RR, Knott TJ, Legros JE, Navaratnam N, Greeve JC, Scott J (September 1991). "Sequence requirements for the editing of apolipoprotein B mRNA". J. Biol. Chem. 266 (25): 16301–16304. doi:10.1016/S0021-9258(18)55296-0. PMID 1885564.
- ^ Driscoll DM, Lakhe-Reddy S, Oleksa LM, Martinez D (December 1993). "Induction of RNA editing at heterologous sites by sequences in apolipoprotein B mRNA". Mol. Cell. Biol. 13 (12): 7288–7294. doi:10.1128/MCB.13.12.7288. PMC 364799. PMID 8246950.
- ^ Greeve J, Navaratnam N, Scott J (July 1991). "Characterization of the apolipoprotein B mRNA editing enzyme: no similarity to the proposed mechanism of RNA editing in kinetoplastid protozoa". Nucleic Acids Res. 19 (13): 3569–3576. doi:10.1093/nar/19.13.3569. PMC 328381. PMID 1649450.
- ^ Richardson N, Navaratnam N, Scott J (November 1998). "Secondary structure for the apolipoprotein B mRNA editing site. Au-binding proteins interact with a stem loop". J. Biol. Chem. 273 (48): 31707–31717. doi:10.1074/jbc.273.48.31707. PMID 9822632.
- ^ Teng B, Verp M, Salomon J, Davidson NO (November 1990). "Apolipoprotein B messenger RNA editing is developmentally regulated and widely expressed in human tissues". J. Biol. Chem. 265 (33): 20616–20620. doi:10.1016/S0021-9258(17)30547-1. PMID 2243107.
- ^ Wu JH, Semenkovich CF, Chen SH, Li WH, Chan L (July 1990). "Apolipoprotein B mRNA editing. Validation of a sensitive assay and developmental biology of RNA editing in the rat". J. Biol. Chem. 265 (21): 12312–12316. doi:10.1016/S0021-9258(19)38347-4. PMID 2373694.
- ^ Glickman RM, Rogers M, Glickman JN (July 1986). "Apolipoprotein B synthesis by human liver and intestine in vitro". Proc. Natl. Acad. Sci. U.S.A. 83 (14): 5296–5300. Bibcode:1986PNAS...83.5296G. doi:10.1073/pnas.83.14.5296. PMC 323938. PMID 3460091.
- ^ Baum CL, Teng BB, Davidson NO (November 1990). "Apolipoprotein B messenger RNA editing in the rat liver. Modulation by fasting and refeeding a high carbohydrate diet". J. Biol. Chem. 265 (31): 19263–19270. doi:10.1016/S0021-9258(17)30653-1. PMID 2229075.
- ^ Lau PP, Cahill DJ, Zhu HJ, Chan L (October 1995). "Ethanol modulates apolipoprotein B mRNA editing in the rat". J. Lipid Res. 36 (10): 2069–2078. doi:10.1016/S0022-2275(20)39192-6. PMID 8576634.
- ^ Chan L, Chang BH, Nakamuta M, Li WH, Smith LC (March 1997). "Apobec-1 and apolipoprotein B mRNA editing". Biochim. Biophys. Acta. 1345 (1): 11–26. doi:10.1016/S0005-2760(96)00156-7. PMID 9084497.
- ^ Chan L (January 1993). "RNA editing: exploring one mode with apolipoprotein B mRNA". BioEssays. 15 (1): 33–41. doi:10.1002/bies.950150106. PMID 8466474. S2CID 314984.
- ^ Tarugi P, Albertazzi L, Nicolini S, Calandra S (March 1990). "Absence of apolipoprotein B-48 in the chick, Gallus domesticus". J. Lipid Res. 31 (3): 417–427. doi:10.1016/S0022-2275(20)43164-5. hdl:11380/742118. PMID 2341807.
- ^ Kumar V, Butcher SJ, Öörni K, Engelhardt P, Heikkonen J, Kaski K, Ala-Korpela M, Kovanen PT (May 2011). "Three-Dimensional cryoEM Reconstruction of Native LDL Particles to 16Å Resolution at Physiological Body Temperature". PLOS ONE. 6 (5): e18841. Bibcode:2011PLoSO...618841K. doi:10.1371/journal.pone.0018841. PMC 3090388. PMID 21573056.
- ^ Knott TJ, Pease RJ, Powell LM, Wallis SC, Rall SC, Innerarity TL, Blackhart B, Taylor WH, Marcel Y, Milne R (1986). "Complete protein sequence and identification of structural domains of human apolipoprotein B". Nature. 323 (6090): 734–738. Bibcode:1986Natur.323..734K. doi:10.1038/323734a0. PMID 3773997. S2CID 536926.
Further reading
[edit]- Mahley RW, Innerarity TL, Rall SC, Weisgraber KH (1985). "Plasma lipoproteins: apolipoprotein structure and function". J. Lipid Res. 25 (12): 1277–1294. doi:10.1016/S0022-2275(20)34443-6. PMID 6099394.
- Itakura H, Matsumoto A (1995). "[Apolipoprotein B]". Nippon Rinsho. 52 (12): 3113–3118. PMID 7853698.
- Chumakova OS, Zateĭshchikov DA, Sidorenko BA (2006). "[Apolipoprotein B: structure, function, gene polymorphism, and relation to atherosclerosis]". Kardiologiia. 45 (6): 43–55. PMID 16007035.
- Ye J (2007). "Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus". PLOS Pathog. 3 (8): e108. doi:10.1371/journal.ppat.0030108. PMC 1959368. PMID 17784784.
External links
[edit]- Database of RNA editing (DARNED).
- Applied Research on Apolipoprotein-B
- Human APOB genome location and APOB gene details page in the UCSC Genome Browser.