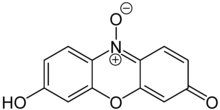
Resazurin
| |
| Names | |
|---|---|
| IUPAC name
7-hydroxy-10-oxidophenoxazin-10-ium-3-one
| |
| Other names
Alamar Blue, Vybrant, UptiBlue, diazo-resorcinol, azoresorcin, resazoin, resazurine
| |
| Identifiers | |
| |
3D model (JSmol)
|
|
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.008.171 |
PubChem CID
|
|
| UNII | |
CompTox Dashboard (EPA)
|
|
| |
| Properties | |
| C12H7NO4 | |
| Molar mass | 229.191 g·mol−1 |
| soluble | |
| Hazards | |
| GHS labelling: | |

| |
| Warning | |
| H315, H319, H335 | |
| P261, P305+P351+P338 | |
| NFPA 704 (fire diamond) | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
Resazurin (7-Hydroxy-3H-phenoxazin-3-one 10-oxide) is a phenoxazine dye that is weakly fluorescent, nontoxic, cell-permeable, and redox‐sensitive.[2][3] Resazurin has a blue to purple color above pH 6.5 and an orange color below pH 3.8.[4] It is used in microbiological, cellular, and enzymatic assays because it can be irreversibly reduced to the pink-colored and highly fluorescent resorufin (7-Hydroxy-3H-phenoxazin-3-one). At circum-neutral pH, resorufin can be detected by visual observation of its pink color or by fluorimetry, with an excitation maximum at 530-570 nm and an emission maximum at 580-590 nm.[5]
When a solution containing resorufin is submitted to reducing conditions (Eh < -110 mV), almost all resorufin is reversibly reduced to the translucid non-fluorescent dihydroresorufin (also known as hydroresorufin) and the solution becomes translucid (the redox potential of the resorufin/dihydroresorufin pair is -51 mV vs. standard hydrogen electrode at pH 7.0). When the Eh of this same solution is increased, dihydroresorufin is oxidized back to resorufin, and this reversible reaction can be used to monitor if the redox potential of a culture medium remains at a sufficiently low level for anaerobic organisms.
| Resazurin (pH indicator) | ||
| below pH 3.8 | above pH 6.5 | |
| 3.8 | ⇌ | 6.5 |
Resazurin solution has one of the highest values known of Kreft's dichromaticity index.[6] This means that it has a large change in perceived color hue when the thickness or concentration of observed sample increases or decreases.
Usually, resazurin is available commercially as the sodium salt.
Cell viability applications
[edit]Resazurin is reduced to resorufin by aerobic respiration of metabolically active cells, and it can be used as an indicator of cell viability. It was first used to quantify bacterial content in milk by Pesch and Simmert in 1929.[7] It can be used to detect the presence of viable cells in mammalian cell cultures.[8] It was introduced commercially initially under Alamar Blue trademark (Trek Diagnostic Systems, Inc), and now also available under other names such as AB assay, Vybrant (Molecular Probes) and UptiBlue (Interchim).
Resazurin based assays show excellent correlation to reference viability assays such as formazan-based assays (MTT/XTT) and tritiated thymidine based techniques.[9][10] The low toxicity makes it suitable for longer studies, and it has been applied for animal cells, bacteria, and fungi [10] for cell culture assays such as cell counting, cell survival, and cell proliferation.[11] In antimicrobial assays, resazurin is commonly utilized to assess the minimum inhibitory concentration (MIC) or minimum bactericidal concentration (MBC) of antimicrobial agents. [12]
To take the place of a standard live/dead assay, resazurin also be multiplexed with chemiluminescent assays, such as cytokine assays, caspase assays to measure apoptosis, or reporter assays to measure a gene or a protein expression.[10]
The irreversible reaction of resazurin to resorufin is proportional to aerobic respiration.[13]
-
Resazurin as a colorimetric assay for cell viability
-
Resazurin used as a fluorescent assay for cell viability - Resazurin does not fluoresce when exposed to green light
-
Resazurin as a fluorescent assay for cell viability - Resorufin fluoresces when exposed to green light
Resazurin reduction test
[edit]Resazurin can be used as one of a series of rapid tests to determine the quality of a milk sample. In this test, resazurin is added as a violet redox dye which turns mauvish-pink due to conversion to resorufin and then to colourless dihydroresorufin. This happens due to lowering of the oxidation-reduction potential in the milk sample caused by presence of bacteria which utilize available oxygen present in the milk for aerobic respiration. The rate of the colour change is used as an index for the number of bacteria present in the milk sample.[14]
Other applications
[edit]
Resazurin is effectively reduced in mitochondria, making it useful also to assess mitochondrial metabolic activity.
Usually, in the presence of NADPH dehydrogenase or NADH dehydrogenase as the enzyme, NADPH or NADH is the reductant that converts resazurin to resorufin. Hence the resazurin/diaphorase/NADPH system can be used to detect NADH, NADPH, or diaphorase level, and any biochemical or enzyme activity that is involved in a biochemical reaction generating NADH or NADPH.[15][16][17][18][19]
Resazurin can be used to assay L-Glutamate, achieving a sensitivity of 2.0 pmol per well in a 96 well plate.[20]
Resazurin can also be used to measure the aerobic biodegradation of organic matter found in effluents.[21]
Resazurin is used to measure the amount of aerobic respiration in streams.[22] Since most aerobic respiration occurs in the stream bed, the conversion of resazurin to resorufin is also a measure of the amount of exchange between the water column and the stream bed.
Synthesis
[edit]Resazurin is prepared by acid-catalyzed condensation between resorcinol and 4-nitrosoresorcinol followed by oxidation of the intermediate with manganese(IV) oxide:
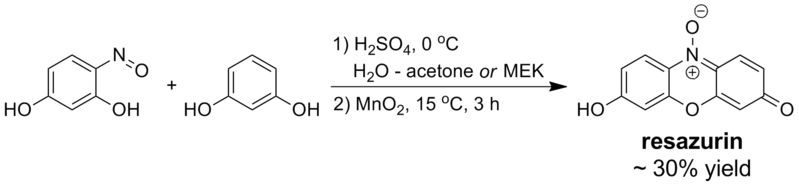
Treatment of the crude reaction product with excess sodium carbonate yields the sodium salt of resazurin, which is typically the commercial form of the dye. Running the condensation step in alcohols is possible but results in lower yields of the product; in pure water or acetic acid, the reaction does not proceed satisfactorily.[23]
Related dyes
[edit]10-acetyl-3,7-dihydroxyphenoxazine (also known as Amplex Red), structurally related to resazurin, reacts with H2O2 in a 1:1 stoichiometry to produce the same by-product resorufin (used in many assays combining for example horseradish peroxidase (HRP), or NADH, NADPH using enzymes).[24]
7-ethoxyresorufin, a compound used as the substrate in the measurement of cytochrome P450 (CYP1A1) induction using the ethoxyresorufin-O-deethylase (EROD) assay system in cell culture and environmental samples, produced in response to exposure to aryl hydrocarbons. The compound is catalysed by the enzyme to produce the same fluorescent product, resorufin.[25][26]
1,3-dichloro-7-hydroxy-9,9-dimethylacridin-2(9H)-one (DDAO dye), a fluorescent dye used for oligonucleotide labeling.[27]
References
[edit]- ^ "Resazurin | C12H7NO4 | ChemSpider".
- ^ Bueno, C.; Villegas, M. L.; Bertolotti, S. G.; Previtali, C. M.; Neumann, M. G.; Encinas, M. V. (2002). "The Excited-State Interaction of Resazurin and Resorufin with Aminesin Aqueous Solutions. Photophysics and Photochemical Reaction". Photochemistry and Photobiology. 76 (4): 385–90. doi:10.1562/0031-8655(2002)0760385TESIOR2.0.CO2. PMID 12405144. S2CID 222102027.
- ^ Haggerty, Roy; Martí, Eugènia; Argerich, Alba; Schiller, Daniel von; Grimm, Nancy B. (2009). "Resazurin as a "smart" tracer for quantifying metabolically active transient storage in stream ecosystems". Journal of Geophysical Research: Biogeosciences. 114 (G3). Bibcode:2009JGRG..114.3014H. doi:10.1029/2008JG000942. hdl:10261/38263. ISSN 2156-2202.
- ^ "Resazurin, sodium salt". www.thomassci.com. Retrieved 2023-12-10.
- ^ Chen, J. L., Steele, T. W., & Stuckey, D. C. (2015). Modeling and application of a rapid fluorescence-based assay for biotoxicity in anaerobic digestion. Environmental science & technology, 49(22), 13463-13471.http://pubs.acs.org/doi/10.1021/acs.est.5b03050
- ^ Kreft, Samo; Kreft, Marko (2009). "Quantification of dichromatism: A characteristic of color in transparent materials". Journal of the Optical Society of America A. 26 (7): 1576–81. Bibcode:2009JOSAA..26.1576K. doi:10.1364/JOSAA.26.001576. PMID 19568292.
- ^ Pesch, K. L.; Simmert, U. (1929). "Combined assays for lactose and galactose by enzymatic reactions". Milchw. Forsch. 8: 551.
- ^ Anoopkumar-Dukie, S; Carey, JB; Conere, T; O'Sullivan, E; Van Pelt, FN; Allshire, A (2005). "Resazurin assay of radiation response in cultured cells". British Journal of Radiology. 78 (934): 945–7. doi:10.1259/bjr/54004230. PMID 16177019.
- ^ Gavanji S, Bakhtari A, Famurewa AC, Othman EM (January 2023). "Cytotoxic Activity of Herbal Medicines as Assessed in Vitro: A Review". Chemistry & Biodiversity. 20 (2): 3–27. doi:10.1002/cbdv.202201098. PMID 36595710. S2CID 255473013.
- ^ a b c UptiBlue viable cell assay technical manual
- ^ Kurin, Elena; Atanasov, Atanas; Donath, Oliver; Heiss, Elke; Dirsch, Verena; Nagy, Milan (2012). "Synergy Study of the Inhibitory Potential of Red Wine Polyphenols on Vascular Smooth Muscle Cell Proliferation". Planta Medica. 78 (8): 772–8. doi:10.1055/s-0031-1298440. PMID 22499559. S2CID 206328057.
- ^ Hossain TJ (July 2023). "Methods for screening and evaluation of antimicrobial activity: a review of protocols, advantages and limitations". Preprint. doi:10.13140/RG.2.2.33198.48965/1.
- ^ González-Pinzón, Ricardo; Haggerty, Roy; Myrold, David D. (2012). "Measuring aerobic respiration in stream ecosystems using the resazurin-resorufin system" (PDF). Journal of Geophysical Research. 117 (G3): G00N06. Bibcode:2012JGRG..117.0N06G. doi:10.1029/2012JG001965.
- ^ Tortorello, Mary Lou, Batt, Carl A. (2 April 2014). Encyclopedia of food microbiology. Academic Press. ISBN 978-0-12-384733-1. OCLC 1049639465.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Shahangian, S.; Ash, K. O.; Rollins, D. E. (1984). "An Enzymatic Method for the Analysis of Formate in Human Plasma". Journal of Analytical Toxicology. 8 (6): 273–6. doi:10.1093/jat/8.6.273. PMID 6549198.
- ^ Hanson, NQ; Freier, EF (1983). "Effect of protein on the determination of total bile acids in serum". Clinical Chemistry. 29 (1): 171–5. doi:10.1093/clinchem/29.1.171. PMID 6571720.
- ^ De Jong, Donald W.; Woodlief, William G. (1977). "Fluorimetric assay of tobacco leaf dehydrogenases with resazurin". Biochimica et Biophysica Acta (BBA) - Enzymology. 484 (2): 249–59. doi:10.1016/0005-2744(77)90081-X. PMID 20957.
- ^ Barnes, Stephen; Spenney, Jerry G. (1980). "Stoichiometry of the nadh-oxidoreductase reaction for dehydrogenase determinations". Clinica Chimica Acta. 107 (3): 149–54. doi:10.1016/0009-8981(80)90442-8. PMID 6893684.
- ^ Winartasaputra, H; Mallet, VN; Kuan, SS; Guilbault, GG (1980). "Fluorometric and colorimetric enzymic determination of triglycerides (triacylglycerols) in serum". Clinical Chemistry. 26 (5): 613–7. doi:10.1093/clinchem/26.5.613. PMID 6894889.
- ^ Chapman, Justin; Zhou, Mingjie (1999). "Microplate-based fluorometric methods for the enzymatic determination of l-glutamate: application in measuring l-glutamate in food samples". Analytica Chimica Acta. 402 (1–2): 47–52. Bibcode:1999AcAC..402...47C. doi:10.1016/S0003-2670(99)00533-4.
- ^ Jouanneau, S.; Recoules, L.; Durand, M.J; Boukabache, A.; Picot, V.; Primault, Y.; Lakel, A.; Sengelin, M.; Barillon, B.; Thouand, G. (2014). "Methods for assessing biochemical oxygen demand (BOD): A review" (PDF). Water Research. 49: 62–82. Bibcode:2014WatRe..49...62J. doi:10.1016/j.watres.2013.10.066. PMID 24316182.
- ^ Haggerty, Roy; Martí, Eugènia; Argerich, Alba; Von Schiller, Daniel; Grimm, Nancy B. (2009). "Resazurin as a 'smart' tracer for quantifying metabolically active transient storage in stream ecosystems". Journal of Geophysical Research. 114 (G3): G03014. Bibcode:2009JGRG..114.3014H. doi:10.1029/2008JG000942. hdl:10261/38263.
- ^ A US 2376283 A, Frank Short Wallace & Peter Oxley, "Dyestuffs suitable for use as indicators", published 1945-05-15, assigned to Boots Pure Drug Co Ltd
- ^ Zhou, M.; Diwu, Z.; Panchuk-Voloshina, N.; Haugland, R.P. (1997). "A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: Applications in detecting the activity of phagocyte NADPH oxidase and other oxidases". Anal Biochem. 253 (2): 162–168. doi:10.1006/abio.1997.2391. PMID 9367498.
- ^ Mohammadi-Bardbori, Afshin (2014). "Assay for quantitative determination of CYP1A1 enzyme activity using 7-Ethoxyresorufin as standard substrate (EROD assay)". Protocol Exchange. 10 (5). Research Square. doi:10.1038/protex.2014.043.
- ^ Chang, Thomas K.H.; Waxman, David J. (2005). "Enzymatic Analysis of cDNA-Expressed Human CYP1A1, CYP1A2, and CYP1B1 With 7-Ethoxyresorufin as Substrate". In Phillips, I.R.; Shephard, E.A. (eds.). Cytochrome P450 Protocols. Methods in Molecular Biology. Vol. 320 (2nd ed.). pp. 85–90. doi:10.1385/1-59259-998-2:85. ISBN 9781588294418. PMID 16719376.
- ^ Dominiak, D.M.; Nielsen, J.L.; Nielsen, P.H. (2010). "Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms". Environmental Microbiology. 13 (3): 710–721. doi:10.1111/j.1462-2920.2010.02375.x. PMID 21118344.




